Journal of Peking University (Health Sciences) ›› 2021, Vol. 53 ›› Issue (2): 420-424. doi: 10.19723/j.issn.1671-167X.2021.02.032
Previous Articles Next Articles
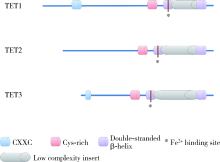
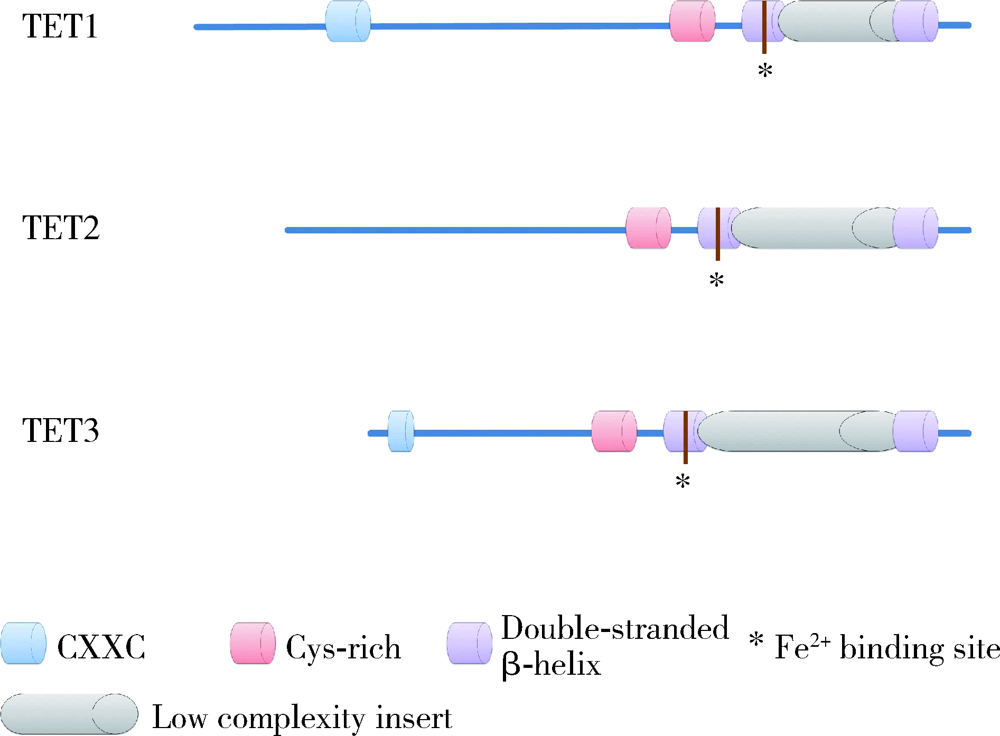
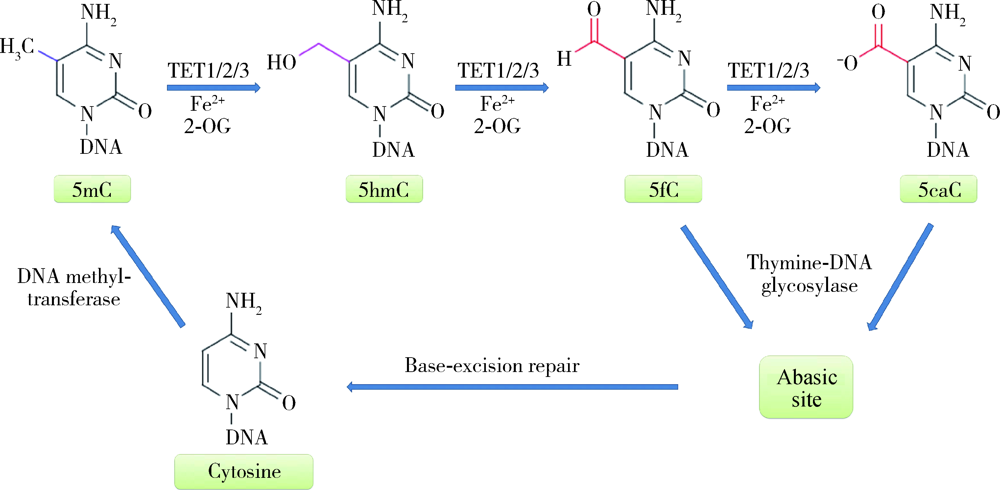
Roles of ten eleven translocation proteins family and 5-hydroxymethylcytosine in epigenetic regulation of stem cells and regenerative medicine
ZHAO Jian-fang1,2,LI Dong1,AN Yang1,Δ( )
)
- 1. Department of Plastic Surgery, Peking University Third Hospital, Beijing 100191, China
2. Department of Plastic Surgery and Burns, Peking University First Hospital, Beijing 100034, China
CLC Number:
- R622
| [1] |
Andrews AJ, Luger K. Nucleosome structure(s) and stability: variations on a theme[J]. Annu Rev Biophys, 2011,40:99-117.
doi: 10.1146/annurev-biophys-042910-155329 pmid: 21332355 |
| [2] |
Bird A. DNA methylation patterns and epigenetic memory[J]. Genes Dev, 2002,16(1):6-21.
doi: 10.1101/gad.947102 pmid: 11782440 |
| [3] |
An J, Rao A, Ko M. TET family dioxygenases and DNA demethylation in stem cells and cancers[J]. Exp Mol Med, 2017,49(4):e323.
doi: 10.1038/emm.2017.5 pmid: 28450733 |
| [4] |
Mizuno H, Tobita M, Uysal AC. Concise review: Adipose-derived stem cells as a novel tool for future regenerative medicine[J]. Stem Cells, 2012,30(5):804-810.
doi: 10.1002/stem.1076 pmid: 22415904 |
| [5] |
Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics[J]. Nat Rev Genet, 2008,9(6):465-476.
doi: 10.1038/nrg2341 pmid: 18463664 |
| [6] |
Kafri T, Ariel M, Brandeis M, et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line[J]. Genes Dev, 1992,6(5):705-714.
doi: 10.1101/gad.6.5.705 pmid: 1577268 |
| [7] |
Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer[J]. Trends Genet, 2014,30(10):464-474.
doi: 10.1016/j.tig.2014.07.005 |
| [8] |
Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1[J]. Science, 2009,324(5929):930-935.
doi: 10.1126/science.1170116 pmid: 19372391 |
| [9] |
Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine[J]. Science, 2011,333(6047):1300-1303.
doi: 10.1126/science.1210597 |
| [10] |
Shi DQ, Ali I, Tang J, et al. New insights into 5hmC DNA modification: generation, distribution and function[J]. Front Genet, 2017,8:100.
pmid: 28769976 |
| [11] |
Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer[J]. Genes Dev, 2016,30(7):733-750.
doi: 10.1101/gad.276568.115 pmid: 27036965 |
| [12] |
Ono R, Taki T, Taketani T, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10, 11)(q22;q23)[J]. Cancer Res, 2002,62(14):4075-4080.
pmid: 12124344 |
| [13] |
Ito S, D’Alessio AC, Taranova OV, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification[J]. Nature, 2010,466(7310):1129-1133.
doi: 10.1038/nature09303 pmid: 20639862 |
| [14] |
Nestor CE, Ottaviano R, Reddington J, et al. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes[J]. Genome Res, 2012,22(3):467-477.
doi: 10.1101/gr.126417.111 |
| [15] |
Wyatt GR, Cohen SS. A new pyrimidine base from bacteriophage nucleic acids[J]. Nature, 1952,170(4338):1072-1073.
doi: 10.1038/1701072a0 pmid: 13013321 |
| [16] |
Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain[J]. Science, 2009,324(5929):929-930.
doi: 10.1126/science.1169786 pmid: 19372393 |
| [17] |
Globisch D, Munzel M, Muller M, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates[J]. PLoS One, 2010,5(12):e15367.
pmid: 21203455 |
| [18] |
Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions[J]. Cell, 2014,156(1/2):45-68.
doi: 10.1016/j.cell.2013.12.019 |
| [19] |
Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation[J]. Nat Genet, 2011,44(1):23-31.
pmid: 22138693 |
| [20] |
Pfaffeneder T, Hackner B, Truss M, et al. The discovery of 5-formylcytosine in embryonic stem cell DNA[J]. Angew Chem Int Ed Engl, 2011,50(31):7008-7012.
doi: 10.1002/anie.201103899 pmid: 21721093 |
| [21] |
Pastor WA, Pape UJ, Huang Y, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells[J]. Nature, 2011,473(7347):394-397.
doi: 10.1038/nature10102 pmid: 21552279 |
| [22] |
Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation[J]. Nature, 2011,473(7347):398-402.
doi: 10.1038/nature10008 pmid: 21460836 |
| [23] |
Lu F, Liu Y, Jiang L, et al. Role of Tet proteins in enhancer activity and telomere elongation[J]. Genes Dev, 2014,28(19):2103-2119.
doi: 10.1101/gad.248005.114 pmid: 25223896 |
| [24] |
Izadpanah R, Trygg C, Patel B, et al. Biologic properties of mesenchymal stem cells derived from bone marrow and adipose tissue[J]. J Cell Biochem, 2006,99(5):1285-1297.
doi: 10.1002/jcb.20904 pmid: 16795045 |
| [25] |
Mizuno H. Adipose-derived stem and stromal cells for cell-based therapy: current status of preclinical studies and clinical trials[J]. Curr Opin Mol Ther, 2010,12(4):442-449.
pmid: 20677095 |
| [26] |
Kim WS, Park BS, Sung JH. Protective role of adipose-derived stem cells and their soluble factors in photoaging[J]. Arch Dermatol Res, 2009,301(5):329-336.
doi: 10.1007/s00403-009-0951-9 pmid: 19396609 |
| [27] |
Yoshimura K, Sato K, Aoi N, et al. Cell-assisted lipotransfer for cosmetic breast augmentation: supportive use of adipose-derived stem/stromal cells[J]. Aesthetic Plast Surg, 2008,32(1):48-55.
doi: 10.1007/s00266-007-9019-4 pmid: 17763894 |
| [28] |
Paduano F, Marrelli M, Amantea M, et al. Adipose tissue as a strategic source of mesenchymal stem cells in bone regeneration: a topical review on the most promising craniomaxillofacial applications[J]. Int J Mol Sci, 2017,18(10):2140.
doi: 10.3390/ijms18102140 |
| [29] |
Rigotti G, Marchi A, Galie M, et al. Clinical treatment of radiotherapy tissue damage by lipoaspirate transplant: a healing process mediated by adipose-derived adult stem cells[J]. Plast Reconstr Surg, 2007,119(5):1409-1422.
doi: 10.1097/01.prs.0000256047.47909.71 pmid: 17415234 |
| [30] |
Akita S, Akino K, Hirano A, et al. Mesenchymal stem cell therapy for cutaneous radiation syndrome[J]. Health Phys, 2010,98(6):858-862.
doi: 10.1097/HP.0b013e3181d3d52c pmid: 20445394 |
| [31] |
Li Y, Zhang W, Gao J, et al. Adipose tissue-derived stem cells suppress hypertrophic scar fibrosis via the p38/MAPK signaling pathway[J]. Stem Cell Res Ther, 2016,7(1):102.
doi: 10.1186/s13287-016-0356-6 pmid: 27484727 |
| [32] |
Seeliger C, Culmes M, Schyschka L, et al. Decrease of global methylation improves significantly hepatic differentiation of Ad-MSCs: possible future application for urea detoxification[J]. Cell Transplant, 2013,22(1):119-131.
pmid: 22507189 |
| [33] |
Tan SJ, Fang JY, Wu Y, et al. Muscle tissue engineering and regeneration through epigenetic reprogramming and scaffold mani-pulation[J]. Sci Rep, 2015,5:16333.
doi: 10.1038/srep16333 pmid: 26548559 |
| [34] |
Zhang RP, Shao JZ, Xiang LX. GADD45A protein plays an essential role in active DNA demethylation during terminal osteogenic differentiation of adipose-derived mesenchymal stem cells[J]. J Biol Chem, 2011,286(47):41083-41094.
doi: 10.1074/jbc.M111.258715 pmid: 21917922 |
| [35] |
Yan X, Ehnert S, Culmes M, et al. 5-azacytidine improves the osteogenic differentiation potential of aged human adipose-derived mesenchymal stem cells by DNA demethylation[J]. PLoS One, 2014,9(6):e90846.
doi: 10.1371/journal.pone.0090846 pmid: 24603866 |
| [36] |
Daniunaite K, Serenaite I, Misgirdaite R, et al. Epigenetic regulation of human adipose-derived stem cells differentiation[J]. Mol Cell Biochem, 2015,410(1/2):111-120.
doi: 10.1007/s11010-015-2543-7 |
| [37] |
Kornicka K, Marycz K, Maredziak M, et al. The effects of the DNA methyltranfserases inhibitor 5-Azacytidine on ageing, oxidative stress and DNA methylation of adipose derived stem cells[J]. J Cell Mol Med, 2017,21(2):387-401.
pmid: 27998022 |
| [38] |
Marycz K, Kornicka K, Irwin-Houston JM, et al. Combination of resveratrol and 5-Azacytidine improves osteogenesis of metabolic syndrome mesenchymal stem cells[J]. J Cell Mol Med, 2018,22(10):4771-4793.
doi: 10.1111/jcmm.13731 pmid: 29999247 |
| [39] |
Kornicka K, Szlapka-Kosarzewska J, Smieszek A, et al. 5-Azacytidine and resveratrol reverse senescence and ageing of adipose stem cells via modulation of mitochondrial dynamics and autophagy[J]. J Cell Mol Med, 2019,23(1):237-259.
doi: 10.1111/jcmm.13914 pmid: 30370650 |
| [40] |
Yoo Y, Park JH, Weigel C, et al. TET-mediated hydroxymethylcytosine at the PPARgamma locus is required for initiation of adipogenic differentiation[J]. Int J Obes (Lond), 2017,41(4):652-659.
doi: 10.1038/ijo.2017.8 |
| [41] |
Fujiki K, Shinoda A, Kano F, et al. PPARgamma-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine[J]. Nat Commun, 2013,4:2262.
doi: 10.1038/ncomms3262 pmid: 23912449 |
| [42] |
Dubois-Chevalier J, Staels B, Lefebvre P, et al. The ubiquitous transcription factor CTCF promotes lineage-specific epigenomic remodeling and establishment of transcriptional networks driving cell differentiation[J]. Nucleus, 2015,6(1):15-18.
pmid: 25565413 |
| [1] | . A novel tissue-engineered bone constructed by using human adipose-derived #br# stem cells and biomimetic calcium phosphate scaffold coprecipitated with #br# bone morphogenetic protein-2 [J]. Journal of Peking University(Health Sciences), 2017, 49(1): 6-015. |
|