Journal of Peking University (Health Sciences) ›› 2021, Vol. 53 ›› Issue (3): 602-607. doi: 10.19723/j.issn.1671-167X.2021.03.028
Previous Articles Next Articles
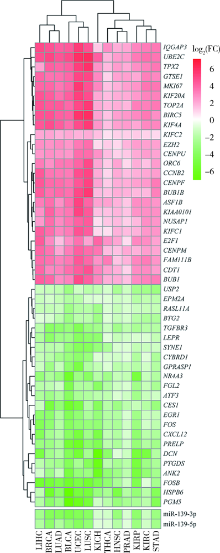
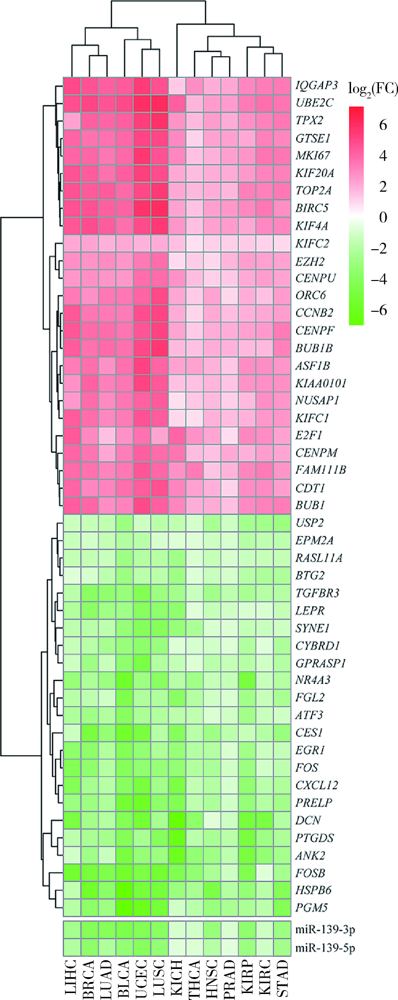
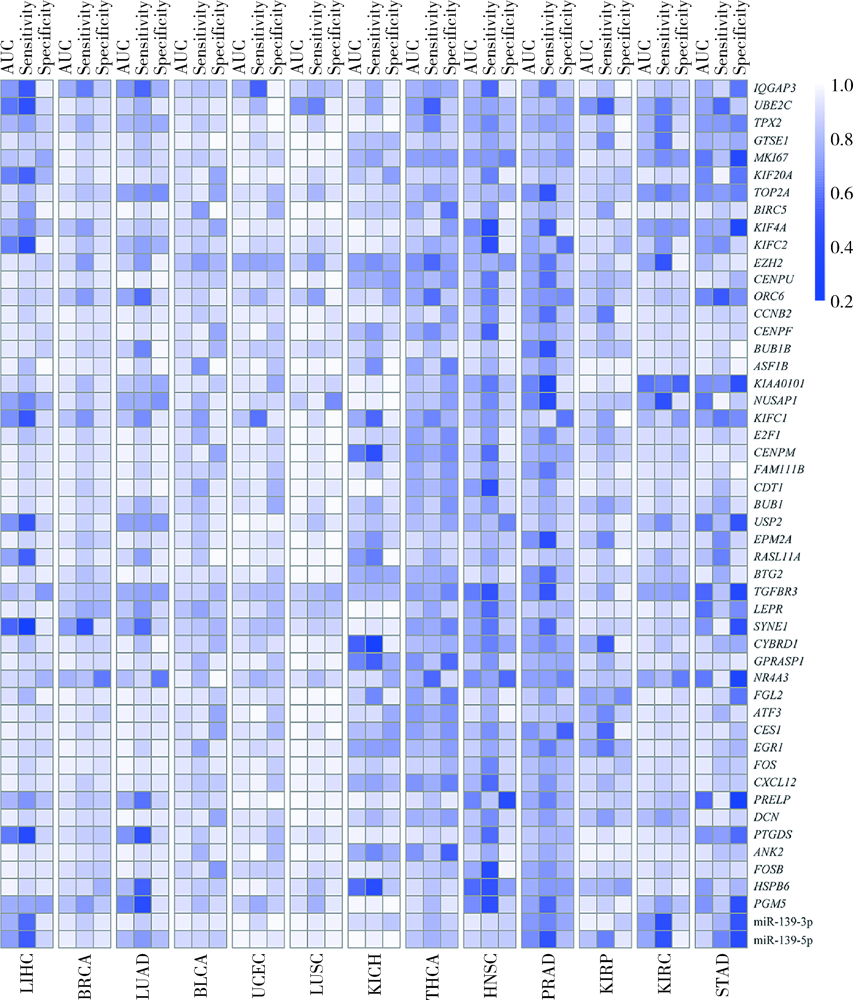
Exploratory screening of potential pan-cancer biomarkers based on The Cancer Genome Atlas database
ZHOU Chuan,MA Xue,XING Yun-kun,LI Lu-di,CHEN Jie,YAO Bi-yun,FU Juan-ling,ZHAO PengΔ( )
)
- Department of Toxicology, Beijing Key Laboratory of Toxicological Research and Risk Assessment for Food Safety, Peking University School of Public Health, Beijing 100191, China
CLC Number:
- R730.43
| [1] |
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018,68(6):394-424.
doi: 10.3322/caac.v68.6 |
| [2] |
Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study[J]. Nat Rev Cancer, 2016,16(8):525-537.
doi: 10.1038/nrc.2016.56 |
| [3] | 王印祥. 泛肿瘤研究和肿瘤免疫研究: 未来抗肿瘤药的发展趋势[J]. 中国药物化学杂志, 2015,25(2):149-152. |
| [4] |
Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration[J]. Nat Genet, 2013,45(10):1134-1140.
doi: 10.1038/ng.2760 |
| [5] | 陈熹, 张峻峰, 曾科, 等. 血清microRNA: 一种非侵入性的肿瘤标志物[J]. 生命科学, 2010,22(7):649-654. |
| [6] |
Li RD, Qu H, Wang SB, et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC[J]. Bioinformatics, 2018,34(14):2515-2517.
doi: 10.1093/bioinformatics/bty124 |
| [7] |
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources[J]. Nat Protoc, 2009,4(1):44-57.
doi: 10.1038/nprot.2008.211 pmid: 19131956 |
| [8] |
Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets[J]. Nucleic Acids Res, 2019,47(D1):D607-D613.
doi: 10.1093/nar/gky1131 |
| [9] |
Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs[J]. Elife, 2015,4:e05005.
doi: 10.7554/eLife.05005 |
| [10] |
Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project[J]. Nat Genet, 2013,45(10):1113-1120.
doi: 10.1038/ng.2764 |
| [11] |
Penault-Llorca F, Radosevic-Robin N. Ki67 assessment in breast cancer: an update[J]. Pathology, 2017,49(2):166-171.
doi: S0031-3025(16)40573-8 pmid: 28065411 |
| [12] |
Yuniati L, Scheijen B, van der Meer LT, et al. Tumor suppressors BTG1 and BTG2: Beyond growth control[J]. J Cell Physiol, 2019,234(5):5379-5389.
doi: 10.1002/jcp.27407 |
| [13] |
Bhattacharjee S, Rajaraman P, Jacobs KB, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits[J]. Am J Hum Genet, 2012,90(5):821-835.
doi: 10.1016/j.ajhg.2012.03.015 |
| [14] |
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation[J]. Cell, 2011,144(5):646-674.
doi: 10.1016/j.cell.2011.02.013 |
| [15] |
Laine A, Westermarck J. Molecular pathways: harnessing E2F1 regulation for prosenescence therapy in p53-defective cancer cells[J]. Clin Cancer Res, 2014,20(14):3644-3650.
doi: 10.1158/1078-0432.CCR-13-1942 pmid: 24788101 |
| [16] |
Xiao YS, Najeeb RM, Ma D, et al. Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms[J]. J Exp Clin Cancer Res, 2019,38(1):458.
doi: 10.1186/s13046-019-1444-0 |
| [17] |
Sun JB, Huang JZ, Lan J, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer[J]. Cancer Cell Int, 2019,19(1):264.
doi: 10.1186/s12935-019-0986-8 |
| [1] | GUO Qian, CHEN Xu-yong, SU Yin. Interleukin-2 signaling pathway regulating molecules in systemic lupus erythematosus [J]. Journal of Peking University(Health Sciences), 2016, 48(6): 1100-1104. |
|
||