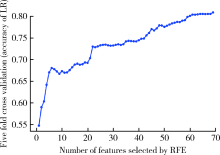
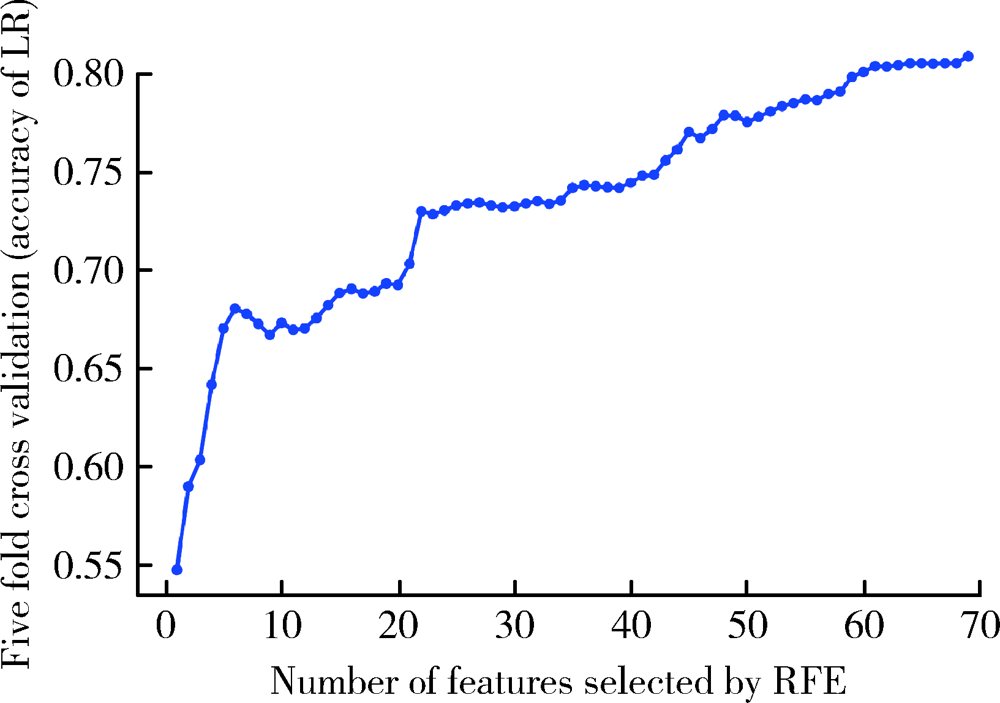
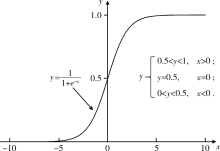
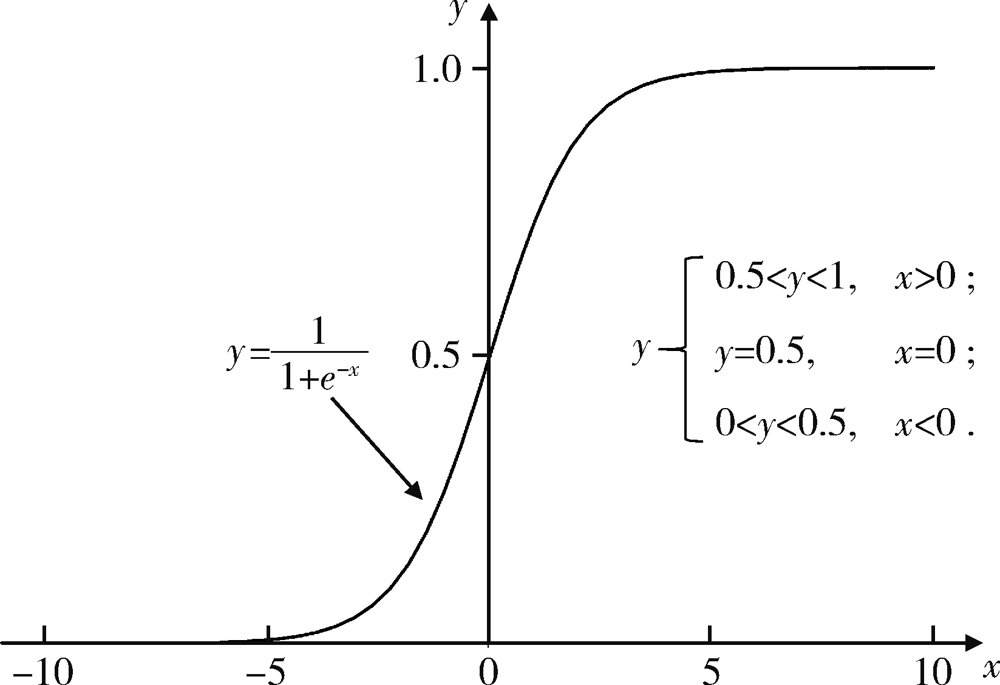
Journal of Peking University (Health Sciences) ›› 2021, Vol. 53 ›› Issue (3): 566-572. doi: 10.19723/j.issn.1671-167X.2021.03.021
Previous Articles Next Articles
Prediction of intensive care unit readmission for critically ill patients based on ensemble learning
LIN Yu1,2,WU Jing-yi3,LIN Ke1,2,HU Yong-hua2,4,KONG Gui-lan1,3,Δ( )
)
- 1. National Institute of Health Data Science, Peking University, Beijing 100191, China
2. Department of Epidemiology and Biostatistics, Peking University School of Public Health, Beijing 100191, China
3. Advanced Institute of Information Technology, Peking University, Hangzhou 311215, China
4. Peking University Medical Informatics Center, Beijing 100191, China
CLC Number:
- R459.7
| [1] |
Halpern NA, Pastores SM. Critical care medicine in the United States 2000-2005: an analysis of bed numbers, occupancy rates, payer mix, and costs[J]. Crit Care Med, 2010,38(1):65-71.
doi: 10.1097/CCM.0b013e3181b090d0 |
| [2] |
Woldhek AL, Rijkenberg S, Bosman RJ, et al. Readmission of ICU patients: A quality indicator?[J]. J Crit Care, 2017,38:328-334.
doi: 10.1016/j.jcrc.2016.12.001 |
| [3] |
Kramer AA, Higgins TL, Zimmerman JE. The association between ICU readmission rate and patient outcomes[J]. Crit Care Med, 2013,41(1):24-33.
doi: 10.1097/CCM.0b013e3182657b8a |
| [4] |
Rosenberg AL, Hofer TP, Hayward RA, et al. Who bounces back? Physiologic and other predictors of intensive care unit readmission[J]. Crit Care Med, 2001,29(3):511-518.
pmid: 11373413 |
| [5] |
Baker DR, Pronovost PJ, Morlock LL, et al. Patient flow variabi-lity and unplanned readmissions to an intensive care unit[J]. Crit Care Med, 2009,37(11):2882-2887.
doi: 10.1097/CCM.0b013e3181b01caf |
| [6] |
Martin LA, Kilpatrick JA, Al-Dulaimi R, et al. Predicting ICU readmission among surgical ICU patients: Development and validation of a clinical nomogram[J]. Surgery, 2019,165(2):373-380.
doi: S0039-6060(18)30429-X pmid: 30170817 |
| [7] |
Lee H, Lim CW, Hong HP, et al. Efficacy of the APACHE Ⅱ score at ICU discharge in predicting post-ICU mortality and ICU readmission in critically ill surgical patients[J]. Anaesth Intensive Care, 2015,43(2):175-186.
doi: 10.1177/0310057X1504300206 |
| [8] |
Fialho AS, Cismondi F, Vieira SM, et al. Data mining using clinical physiology at discharge to predict ICU readmissions[J]. Expert Syst Appl, 2012,39(18):13158-13165.
doi: 10.1016/j.eswa.2012.05.086 |
| [9] |
Desautels T, Das R, Calvert J, et al. Prediction of early unplanned intensive care unit readmission in a UK tertiary care hospital: a cross-sectional machine learning approach[J]. BMJ Open, 2017,7(9):e017199.
doi: 10.1136/bmjopen-2017-017199 |
| [10] |
Hosni M, Abnane I, Idri A, et al. Reviewing ensemble classification methods in breast cancer[J]. Comput Methods Programs Biomed, 2019,177:89-112.
doi: 10.1016/j.cmpb.2019.05.019 |
| [11] | Liu Y, Gu Y, Nguyen JC, et al. Symptom severity classification with gradient tree boosting[J]. J Biomed Inform, 2017,75S:S105-S111. |
| [12] |
Johnson AE, Pollard TJ, Shen L, et al. MIMIC-Ⅲ, a freely accessible critical care database[J]. Sci Data, 2016,3:160035.
doi: 10.1038/sdata.2016.35 |
| [13] |
Austin SR, Wong YN, Uzzo RG, et al. Why summary comorbidity measures such as the Charlson comorbidity index and Elixhauser score work[J]. Med Care, 2015,53(9):E65-E72.
doi: 10.1097/MLR.0b013e318297429c |
| [14] |
Oakes DF, Borges IN, Forgiarini Junior LA, et al. Assessment of ICU readmission risk with the stability and workload index for transfer score[J]. J Bras Pneumol, 2014,40(1):73-76.
doi: 10.1590/S1806-37132014000100011 |
| [15] |
Xue Y, Klabjan D, Luo Y. Predicting ICU readmission using grouped physiological and medication trends[J]. Artif Intell Med, 2019,95:27-37.
doi: S0933-3657(17)30648-6 pmid: 30213670 |
| [16] |
He HB, Garcia EA. Learning from imbalanced data[J]. IEEE T Knowl Data En, 2009,21(9):1263-1284.
doi: 10.1109/TKDE.2008.239 |
| [17] |
Rahman R, Matlock K, Ghosh S, et al. Heterogeneity aware random forest for drug sensitivity prediction[J]. Sci Rep, 2017,7(1):11347.
doi: 10.1038/s41598-017-11665-4 |
| [18] |
Hu J. Automated detection of driver fatigue based on AdaBoost classifier with EEG signals[J]. Front Comput Neurosci, 2017,11:72.
doi: 10.3389/fncom.2017.00072 |
| [19] |
Friedman JH. Greedy function approximation: A gradient boosting machine[J]. Ann Stat, 2001,29(5):1189-1232.
doi: 10.1214/aos/1013203450 |
| [20] | Mani I, Zhang I. kNN approach to unbalanced data distributions: a case study involving information extraction[C]// ICML 2003 Workshop on Learning from Imbalanced Datasets, August 21-24, 2003. Washington, D.C.: ICML, 2003. |
| [1] | WU Jing-yi,LIN Yu,LIN Ke,HU Yong-hua,KONG Gui-lan. Predicting prolonged length of intensive care unit stay via machine learning [J]. Journal of Peking University (Health Sciences), 2021, 53(6): 1163-1170. |
| [2] | Xue-hua ZHU,Ming-yu YANG,Hai-zhui XIA,Wei HE,Zhi-ying ZHANG,Yu-qing LIU,Chun-lei XIAO,Lu-lin MA,Jian LU. Application of machine learning models in predicting early stone-free rate after flexible ureteroscopic lithotripsy for renal stones [J]. Journal of Peking University(Health Sciences), 2019, 51(4): 653-659. |
| [3] | YANG Cheng, ZHANG Yu-qi, TANG Xun, GAO Pei, WEI Chen-lu, HU Yong-hua. Retrospective cohort study for the impact on readmission of patients with ischemic stroke after treatment of aspirin plus clopidogrel or aspirin mono-therapy [J]. Journal of Peking University(Health Sciences), 2016, 48(3): 442-447. |
|
||