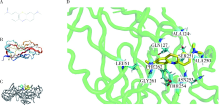
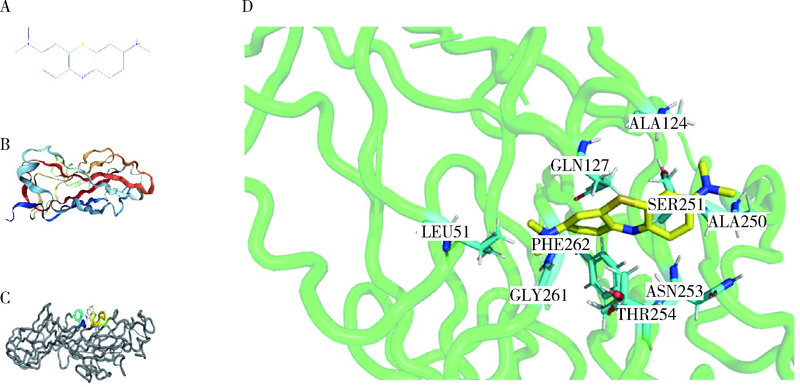
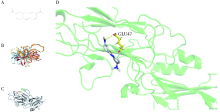
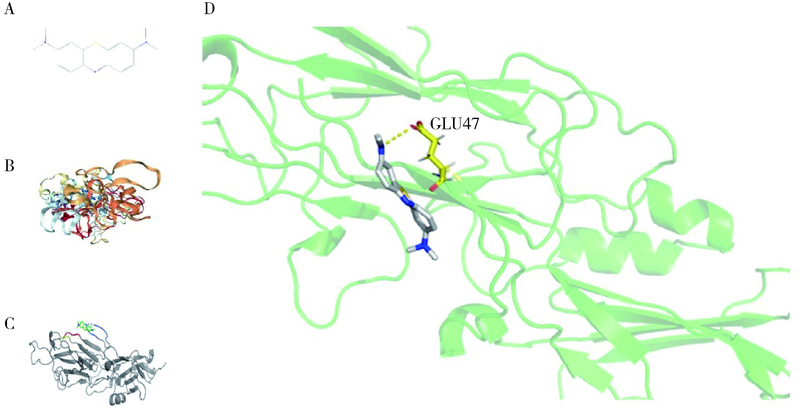
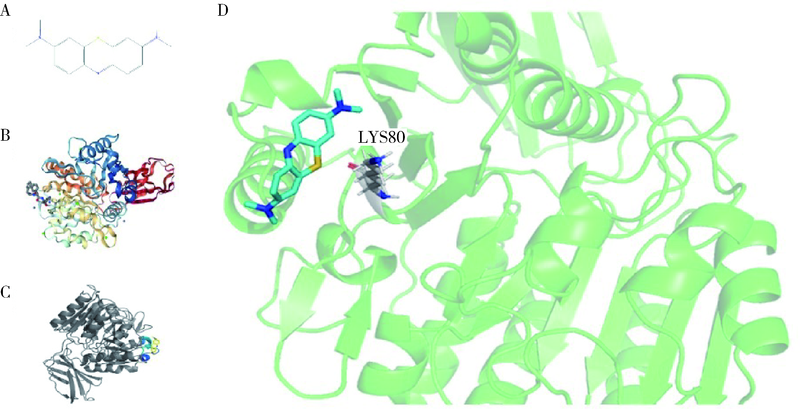
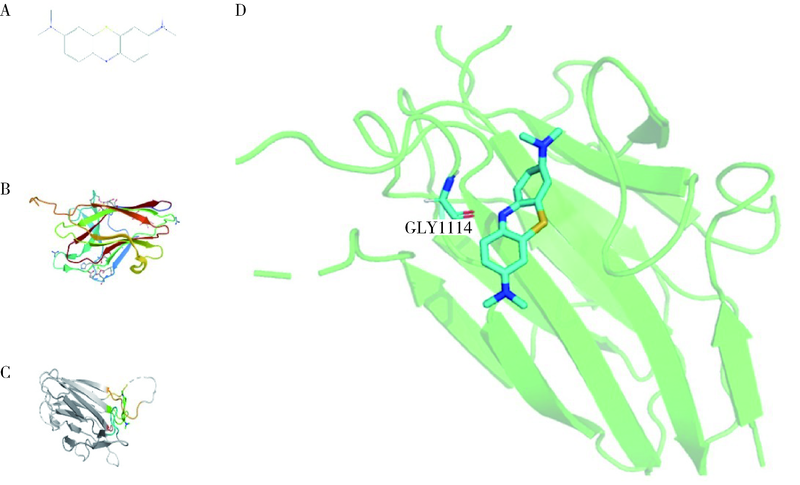
Journal of Peking University (Health Sciences) ›› 2022, Vol. 54 ›› Issue (1): 23-30. doi: 10.19723/j.issn.1671-167X.2022.01.005
Previous Articles Next Articles
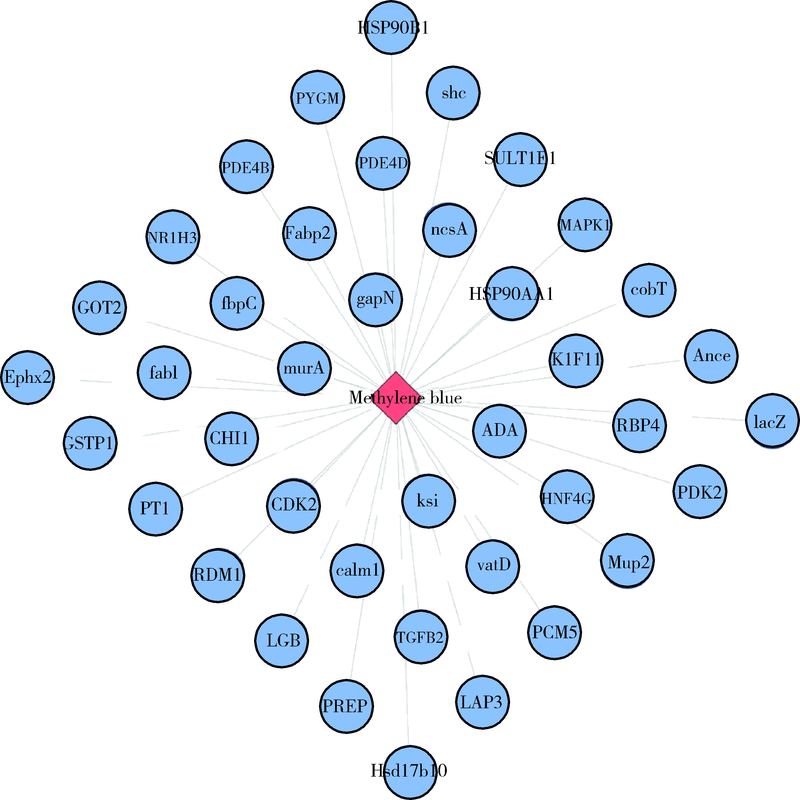
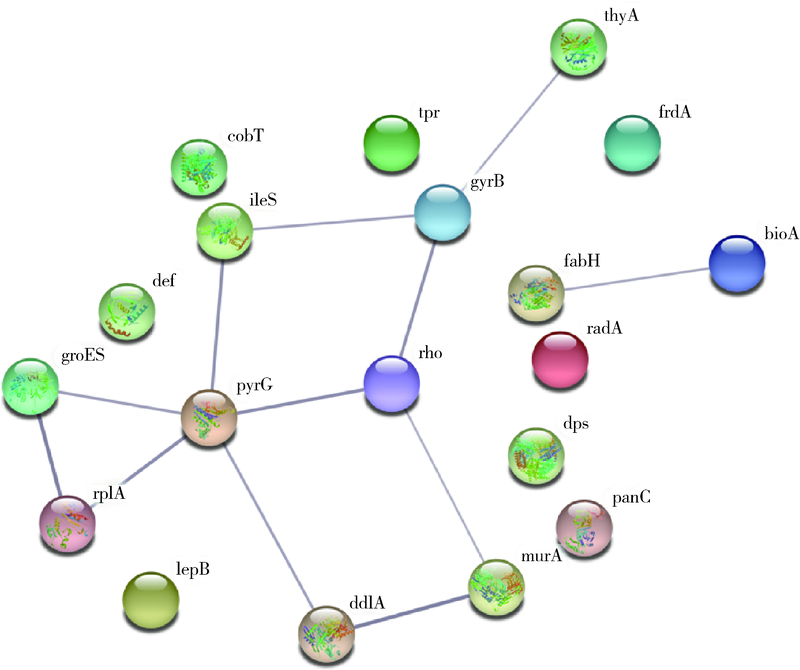
Computer simulation of molecular docking between methylene blue and some proteins of Porphyromonas gingivalis
YUAN Lin-tian1,2,MA Li-sha2,LIU Run-yuan3,QI wei1,2,ZHANG Lu-dan2,4,WANG Gui-yan2,5,WANG Yu-guang2,△( )
)
- 1. Department of General Medicine, Peking University School and Hospital of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Laboratory for Digital and Material Technology of Stomatology & Beijing Key Laboratory of Digital Stomatology, Beijing 100081, China
2. Center for Digital Dentistry, Peking University School and Hospital of Stomatology, Beijing 100081, China
3. Department of Endodontics, College of Stomatology, Dalian Medical University, Dalian 116044, Liaoning, China
4. First Clinical Division, Peking University School and Hospital of Stomatology, Beijing 100081, China
5. Department of Pediatric Dentistry, Peking University School and Hospital of Stomatology, Beijing 100081, China
CLC Number:
- R781.4
| [1] |
Aoyama N, Suzuki JI, Kobayashi N, et al. Associations among tooth loss, systemic inflammation and antibody titers to periodontal pathogens in Japanese patients with cardiovascular disease[J]. J Periodontal Res, 2018, 53(1):117-122.
doi: 10.1111/jre.12494 pmid: 29139559 |
| [2] | Papapanou PN, Sanz M, Budunneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions[J]. J Periodontol, 2018, 89(Suppl 1):173-182. |
| [3] |
Yan X, Lu H, Zhang L, et al. A three-year study on periodontal microorganisms of short locking-taper implants and adjacent teeth in patients with history of periodontitis[J]. J Dent, 2020, 95:103299.
doi: 10.1016/j.jdent.2020.103299 |
| [4] | 梁雨晴, 董秤均, 伍文彬. 牙龈卟啉单胞菌与阿尔茨海默病的相关性研究进展[J]. 中华神经医学杂志, 2020, 19(5):525-527. |
| [5] | 王春萌, 洪丽华, 张志民, 等. 牙龈卟啉单胞菌在消化系统恶性肿瘤中的作用及机制[J]. 华西口腔医学杂志, 2019, 37(5):521-526. |
| [6] |
Kou Y, Inaba H, Kato T, et al. Inflammatory responses of gingival epithelial cells stimulated with Porphyromonas gingivalis vesicles are inhibited by hop-associated polyphenols[J]. J Periodontol, 2008, 79(1):174-180.
doi: 10.1902/jop.2008.070364 |
| [7] |
Ikal R, Hasegawa Y, Izumigawa M, et al. Mfa4, an accessory protein of Mfa1 fimbriae, modulates fimbrial biogenesis, cell auto-aggregation, and biofilm formation in Porphyromonas gingivalis[J]. PLoS One, 2015, 10(10):e0129454
doi: 10.1371/journal.pone.0129454 |
| [8] |
Kato T, Kawai S, Nakano K, et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype[J]. Cell Microbiol, 2007, 9(3):753-765.
doi: 10.1111/cmi.2007.9.issue-3 |
| [9] |
Cieplik F, Deng D, Crielaard W, et al. Antimicrobial photodynamic therapy: what we know and what we don’t[J]. Crit Rev Microbiol, 2018, 44(5):571-589.
doi: 10.1080/1040841X.2018.1467876 pmid: 29749263 |
| [10] |
Kwiatkowski S, Knap B, Prielaard D, et al. Photodynamic therapy mechanisms, photosensitizers and combinations[J]. Biomed Pharmacother, 2018, 106:1098-1107.
doi: S0753-3322(18)34161-1 pmid: 30119176 |
| [11] |
Malik Z, Ladan H, Nitzan Y. Photodynamic inactivation of Gram-negative bacteria: problems and possible solutions[J]. J Photochem Photobiol B, 1992, 14(3):262-266.
doi: 10.1016/1011-1344(92)85104-3 |
| [12] |
Nitzan Y, Gutterman M, Malik Z, et al. Inactivation of gram-negative bacteria by photosensitized porphyrins[J]. Photochem Photobiol, 1992, 55(1):89-96.
pmid: 1534909 |
| [13] |
Malik Z, Hanania J, Nitzan Y. Bactericidal effects of photoactivated porphyrins: an alternative approach to antimicrobial drugs[J]. J Photochem Photobiol B, 1990, 5(3/4):281-293.
doi: 10.1016/1011-1344(90)85044-W |
| [14] |
Wang KK, Finlay JC, Busch TM, et al. Explicit dosimetry for photodynamic therapy: macroscopic singlet oxygen modeling[J]. J Biophotonics, 2010, 3(5/6):304-318.
doi: 10.1002/jbio.v3:5/6 |
| [15] |
Kuimove MK, Yahioglu G, Ogilby PR. Singlet oxygen in a cell: spatially dependent lifetimes and quenching rate constants[J]. J Am Chem Soc, 2009, 131(1):332-340.
doi: 10.1021/ja807484b |
| [16] |
Redmond RW, Kochevar IE. Spatially resolved cellular responses to singlet oxygen[J]. Photochem Photobiol, 2006, 82(5):1178-1186.
pmid: 16740059 |
| [17] |
Pourhajibagher M, Bahador A. Gene expression profiling of fimA gene encoding fimbriae among clinical isolates of Porphyromonas gingivalis in response to photo-activated disinfection therapy[J]. Photodiagnosis Photodyn Ther, 2017, 20:1-5.
doi: S1572-1000(17)30343-5 pmid: 28797828 |
| [18] |
Pourhajibagher M, Bahador A. Evaluation of the crystal structure of a fimbrillin (FimA) from Porphyromonas gingivalis as a therapeutic target for photo-activated disinfection with toluidine blue O[J]. Photodiagnosis Photodyn Ther, 2017, 17:98-102.
doi: S1572-1000(16)30199-5 pmid: 27890593 |
| [19] |
Pourhajibagher M, Bahador A. In silico identification of a therapeutic target for photo-activated disinfection with indocyanine green: modeling and virtual screening analysis of Arg-gingipain from Porphyromonas gingivalis[J]. Photodiagnosis Photodyn Ther, 2017, 18:149-154.
doi: S1572-1000(17)30006-6 pmid: 28254551 |
| [20] |
Forli S, Huey R, Pique ME, et al. Computational protein-ligand docking and virtual drug screening with the AutoDock suite[J]. Nat Protoc, 2016, 11(5):905-919.
doi: 10.1038/nprot.2016.051 |
| [21] |
Ongarora BG, Fontenot KR, HU X, et al. Phthalocyanine-peptide conjugates for epidermal growth factor receptor targeting[J]. J Med Chem, 2012, 55(8):3725-3738.
doi: 10.1021/jm201544y pmid: 22468711 |
| [22] |
Shanmugaraj K, Anandakumar S, Ilanchelian M. Unraveling the binding interaction of Toluidine blue O with bovine hemoglobin: a multi spectroscopic and molecular modeling approach[J]. Rsc Advances, 2015, 5(6):3930-3940.
doi: 10.1039/C4RA11136B |
| [23] | Tsvetkov VB, Soloveva AB, Melik-Nubarov NS. Computer modeling of the complexes of Chlorin e6 with amphiphilic polymers[J]. Phys Chem Chem Phy, 2014, 16(22):10903-10913. |
| [24] |
Zhang Y, Wan Y, Chen Y, et al. Ultrasound-enhanced chemo-photodynamic combination therapy by using albumin “nanoglue”-based nanotheranostics[J]. Acs Nano, 2020, 14(5):5560-5569.
doi: 10.1021/acsnano.9b09827 |
| [25] |
Kandoussi I, Lakhlili W, Taoufik J, et al. Docking analysis of verteporfin with YAP WW domain[J]. Bioinformation, 2017, 13(7):237-240.
doi: 10.6026/97320630013237 pmid: 28943729 |
| [26] |
Alves E, Faustino MA, Neves MG, et al. An insight on bacterial cellular targets of photodynamic inactivation[J]. Future Med Chem, 2014, 6(2):141-164.
doi: 10.4155/fmc.13.211 |
| [27] |
Nagano K. FimA fimbriae of the periodontal disease-associated bacterium Porphyromonas gingivalis[J]. Yakugaku Zasshi, 2013, 133(9):963-974.
pmid: 23995804 |
| [28] |
Xu Q, Shoji M, Shibata S, et al. A distinct type of pilus from the human microbiome[J]. Cell, 2016, 165(3):690-703.
doi: 10.1016/j.cell.2016.03.016 |
| [29] |
Zhou XY, Gao JL, Hunter N, et al. Sequence-independent processing site of the C-terminal domain (CTD) influences maturation of the RgpB protease from Porphyromonas gingivalis[J]. Mol Microbiol, 2013, 89(5):903-917.
doi: 10.1111/mmi.2013.89.issue-5 |
| [30] | 尧晨光, 奚彩丽, 朱祥, 等. EV71 3C蛋白酶表达纯化、活性分析及与抑制剂模拟对接研究[J]. 中国病原生物学杂志, 2017, 12(8):722-726. |
| [31] | 刘福和, 陈少军, 倪文娟. 川芎中抗血栓活性成分的计算机虚拟筛选研究[J]. 中国药房, 2017, 28(16):2182-2186. |
| [1] | Yu-xiao WU,Yi-fan KANG,Qian-ying MAO,Zi-meng LI,Xiao-feng SHAN,Zhi-gang CAI. Application of methylene blue near-infrared fluorescence imaging for oral sentinel lymph node mapping in rats [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 684-688. |
| [2] | CAO Ze,WANG Le-tong,LIU Zhen-ming. Homologous modeling and binding ability analysis of Spike protein after point mutation of severe acute respiratory syndrome coronavirus 2 to receptor proteins and potential antiviral drugs [J]. Journal of Peking University (Health Sciences), 2021, 53(1): 150-158. |
| [3] | Yan XUAN,Yu CAI,Xiao-xuan WANG,Qiao SHI,Li-xin QIU,Qing-xian LUAN. Effect of Porphyromonas gingivalis infection on atherosclerosis in apolipoprotein-E knockout mice [J]. Journal of Peking University (Health Sciences), 2020, 52(4): 743-749. |
|
||