Journal of Peking University (Health Sciences) ›› 2022, Vol. 54 ›› Issue (3): 387-393. doi: 10.19723/j.issn.1671-167X.2022.03.001
Exploring the association between de novo mutations and non-syndromic cleft lip with or without palate based on whole exome sequencing of case-parent trios
Xi CHEN1,Si-yue WANG1,En-ci XUE1,Xue-heng WANG1,He-xiang PENG1,Meng FAN1,Meng-ying WANG1,Yi-qun WU1,Xue-ying QIN1,Jing LI1,Tao WU1,*( ),Hong-ping ZHU2,Jing LI3,Zhi-bo ZHOU2,Da-fang CHEN1,Yong-hua HU1
),Hong-ping ZHU2,Jing LI3,Zhi-bo ZHOU2,Da-fang CHEN1,Yong-hua HU1
- 1. Department of Epidemiology and Biostatistics, Peking University School of Public Health, Beijing 100191, China
2. Department of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology & National Center of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Research Center of Oral Biomaterials and Digital Medical Devices & Beijing Key Laboratory of Digital Stomatology & NHC Research Center of Engineering and Technology for Computerized Dentistry & NMPA Key Laboratory for Dental Materials, Beijing 100081, China
3. Department of Pediatric Dentistry, Peking University School and Hospital of Stomatology & National Center of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Research Center of Oral Biomaterials and Digital Medical Devices & Beijing Key Laboratory of Digital Stomatology & NHC Research Center of Engineering and Technology for Computerized Dentistry & NMPA Key Laboratory for Dental Materials, Beijing 100081, China
CLC Number:
- R181.3+3
| 1 |
Worley ML , Patel KG , Kilpatrick LA . Cleft lip and palate[J]. Clin Perinatol, 2018, 45 (4): 661- 678.
doi: 10.1016/j.clp.2018.07.006 |
| 2 |
Beaty TH , Murray JC , Marazita ML , et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4[J]. Nat Genet, 2010, 42 (6): 525- 529.
doi: 10.1038/ng.580 |
| 3 | Nasreddine G, El Hajj J, Ghassibe-Sabbagh M. Orofacial clefts embryology, classification, epidemiology, and genetics[J/OL]. Mutat Res Rev Mutat Res, 2021, 787: 108373(2021-02-28)[2022-02-01]. https://pubmed.ncbi.nlm.nih.gov/34083042/. |
| 4 |
van Rooij IA , Ludwig KU , Welzenbach J , et al. Non-syndromic cleft lip with or without cleft palate: Genome-wide association study in Europeans identifies a suggestive risk locus at 16p12.1 and supports as a clefting susceptibility gene[J]. Genes (Basel), 2019, 10 (12): 1023.
doi: 10.3390/genes10121023 |
| 5 |
Bishop MR , Diaz Perez KK , Sun M , et al. Genome-wide enrichment of de novo coding mutations in orofacial cleft trios[J]. Am J Hum Genet, 2020, 107 (1): 124- 136.
doi: 10.1016/j.ajhg.2020.05.018 |
| 6 |
Jin ZB , Li Z , Liu Z , et al. Identification of de novo germline mutations and causal genes for sporadic diseases using trio-based whole-exome/genome sequencing[J]. Biol Rev Camb Philos Soc, 2018, 93 (2): 1014- 1031.
doi: 10.1111/brv.12383 |
| 7 |
Conrad DF , Keebler JE , DePristo MA , et al. Variation in genome-wide mutation rates within and between human families[J]. Nat Genet, 2011, 43 (7): 712- 714.
doi: 10.1038/ng.862 |
| 8 |
Veltman JA , Brunner HG . De novo mutations in human genetic disease[J]. Nat Rev Genet, 2012, 13 (8): 565- 575.
doi: 10.1038/nrg3241 |
| 9 |
Coe BP , Stessman HAF , Sulovari A , et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity[J]. Nat Genet, 2019, 51 (1): 106- 116.
doi: 10.1038/s41588-018-0288-4 |
| 10 |
Mitra I , Huang B , Mousavi N , et al. Patterns of de novo tandem repeat mutations and their role in autism[J]. Nature, 2021, 589 (7841): 246- 250.
doi: 10.1038/s41586-020-03078-7 |
| 11 |
Jin SC , Homsy J , Zaidi S , et al. Contribution of rare inherited and de novo variants in 2 871 congenital heart disease probands[J]. Nat Genet, 2017, 49 (11): 1593- 1601.
doi: 10.1038/ng.3970 |
| 12 |
Watkins WS , Hernandez EJ , Wesolowski S , et al. De novo and recessive forms of congenital heart disease have distinct genetic and phenotypic landscapes[J]. Nat Commun, 2019, 10 (1): 4722.
doi: 10.1038/s41467-019-12582-y |
| 13 | Ware JS, Samocha KE, Homsy J, et al. Interpreting de novo variation in human disease using denovolyzeR[J/OL]. Curr Protoc Hum Genet, 2015, 87: 7.25.1 -7.25.15(2015-08-06)[2022-02-01]. https://pubmed.ncbi.nlm.nih.gov/26439716/. |
| 14 |
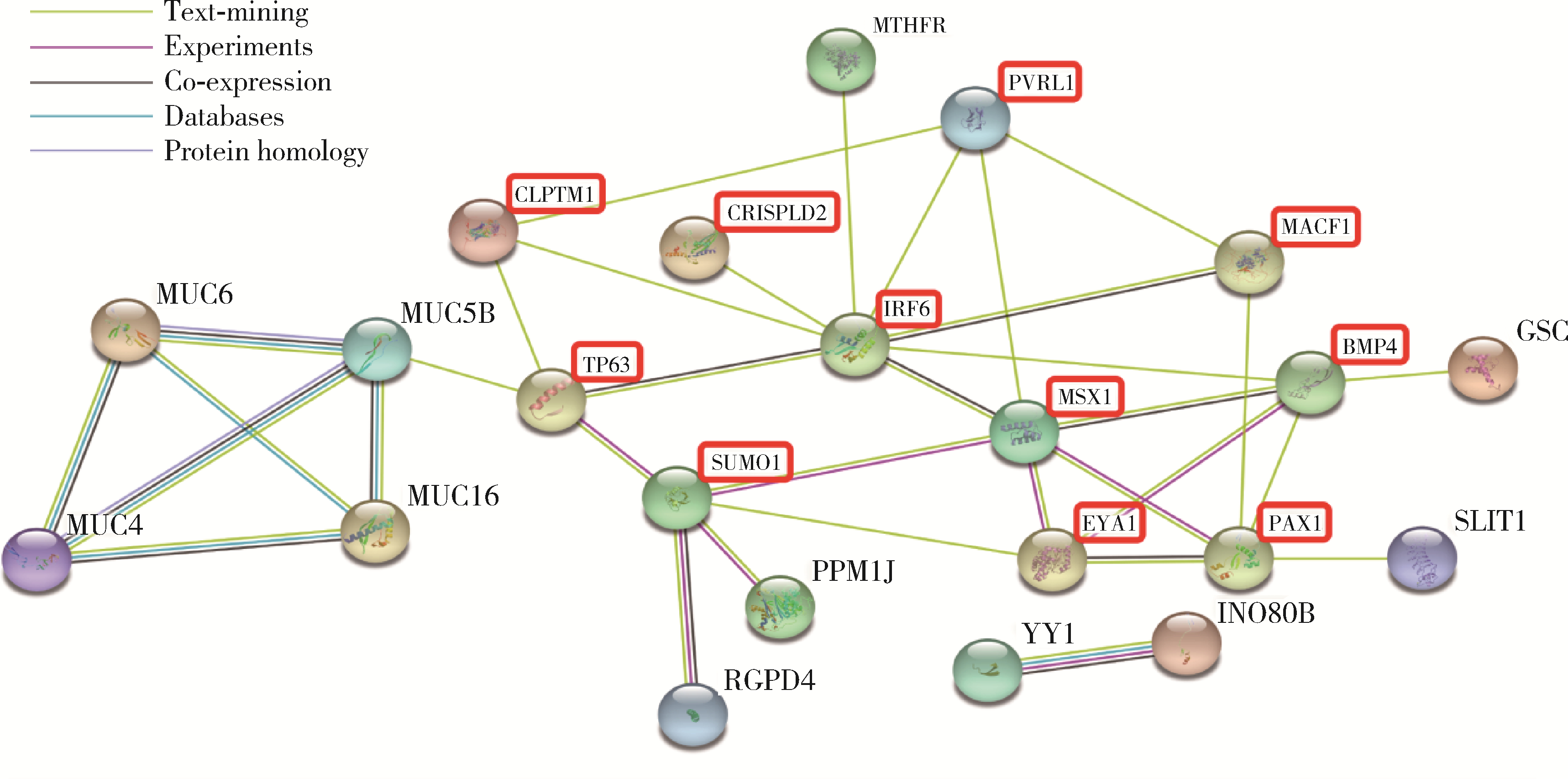
Szklarczyk D , Gable AL , Nastou KC , et al. The STRING database in 2021:Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets[J]. Nucleic Acids Res, 2021, 49 (D1): D605- D612.
doi: 10.1093/nar/gkaa1074 |
| 15 |
Saleem K , Zaib T , Sun W , et al. Assessment of candidate genes and genetic heterogeneity in human non syndromic orofacial clefts specifically non syndromic cleft lip with or without palate[J]. Heliyon, 2019, 5 (12): e03019.
doi: 10.1016/j.heliyon.2019.e03019 |
| 16 |
Vezain M , Lecuyer M , Rubio M , et al. A de novo variant in ADGRL2 suggests a novel mechanism underlying the previously undescribed association of extreme microcephaly with severely reduced sulcation and rhombencephalosynapsis[J]. Acta Neuropathol Commun, 2018, 6 (1): 109.
doi: 10.1186/s40478-018-0610-5 |
| 17 |
Shao R , Liu J , Yan G , et al. Cdh1 regulates craniofacial development via APC-dependent ubiquitination and activation of Goosecoid[J]. Cell Res, 2016, 26 (6): 699- 712.
doi: 10.1038/cr.2016.51 |
| 18 |
Hamann J , Aust G , Araç D , et al. International union of basic and clinical pharmacology. XCIV. Adhesion G protein-coupled receptors[J]. Pharmacol Rev, 2015, 67 (2): 338- 367.
doi: 10.1124/pr.114.009647 |
| 19 |
Sevastre AS , Buzatu IM , Baloi C , et al. ELTD1:An emerging silent actor in cancer drama play[J]. Int J Mol Sci, 2021, 22 (10): 5151.
doi: 10.3390/ijms22105151 |
| 20 |
Lek M , Karczewski KJ , Minikel EV , et al. Analysis of protein-coding genetic variation in 60, 706 humans[J]. Nature, 2016, 536 (7616): 285- 291.
doi: 10.1038/nature19057 |
| 21 |
Huang N , Lee I , Marcotte EM , et al. Characterising and predicting haploinsufficiency in the human genome[J]. PLoS Genet, 2010, 6 (10): e1001154.
doi: 10.1371/journal.pgen.1001154 |
| 22 |
Hiramatsu H , Tadokoro S , Nakanishi M , et al. Latrotoxin-induced exocytosis in mast cells transfected with latrophilin[J]. Toxicon, 2010, 56 (8): 1372- 1380.
doi: 10.1016/j.toxicon.2010.08.002 |
| 23 |
Zepeda-Mendoza CJ , Bardon A , Kammin T , et al. Phenotypic interpretation of complex chromosomal rearrangements informed by nucleotide-level resolution and structural organization of chromatin[J]. Eur J Hum Genet, 2018, 26 (3): 374- 381.
doi: 10.1038/s41431-017-0068-0 |
| 24 |
Passi GR , Bhatnagar S . Rhombencephalosynapsis[J]. Pediatr Neurol, 2015, 52 (6): 651- 652.
doi: 10.1016/j.pediatrneurol.2015.02.005 |
| 25 |
Birnbaum S , Ludwig KU , Reutter H , et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24[J]. Nat Genet, 2009, 41 (4): 473- 477.
doi: 10.1038/ng.333 |
| 26 |
Mangold E , Ludwig KU , Birnbaum S , et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate[J]. Nat Genet, 2010, 42 (1): 24- 26.
doi: 10.1038/ng.506 |
| 27 |
Beaty TH , Taub MA , Scott AF , et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study[J]. Hum Genet, 2013, 132 (7): 771- 781.
doi: 10.1007/s00439-013-1283-6 |
| 28 |
Parry DA , Logan CV , Stegmann AP , et al. SAMS, a syndrome of short stature, auditory-canal atresia, mandibular hypoplasia, and skeletal abnormalities is a unique neurocristopathy caused by mutations in Goosecoid[J]. Am J Hum Genet, 2013, 93 (6): 1135- 1142.
doi: 10.1016/j.ajhg.2013.10.027 |
| 29 |
Ulmer B , Tingler M , Kurz S , et al. A novel role of the organizer gene Goosecoid as an inhibitor of Wnt/PCP-mediated convergent extension in Xenopus and mouse[J]. Sci Rep, 2017, 7, 43010.
doi: 10.1038/srep43010 |
| 30 |
Yu Y , Zuo X , He M , et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity[J]. Nat Commun, 2017, 8, 14364.
doi: 10.1038/ncomms14364 |
| 31 |
Kalisz M , Winzi M , Bisgaard HC , et al. EVEN-SKIPPED HOMEOBOX 1 controls human ES cell differentiation by directly repressing GOOSECOID expression[J]. Dev Biol, 2012, 362 (1): 94- 103.
doi: 10.1016/j.ydbio.2011.11.017 |
| 32 |
Rivera-Pérez JA , Mallo M , Gendron-Maguire M , et al. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development[J]. Development, 1995, 121 (9): 3005- 3012.
doi: 10.1242/dev.121.9.3005 |
| 33 |
Yamada G , Mansouri A , Torres M , et al. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death[J]. Development, 1995, 121 (9): 2917- 2922.
doi: 10.1242/dev.121.9.2917 |
| 34 |
Feitosa NM , Zhang J , Carney TJ , et al. Hemicentin 2 and fibulin 1 are required for epidermal-dermal junction formation and fin mesenchymal cell migration during zebrafish development[J]. Dev Biol, 2012, 369 (2): 235- 248.
doi: 10.1016/j.ydbio.2012.06.023 |
| [1] | Hong-chen ZHENG,En-ci XUE,Xue-heng WANG,Xi CHEN,Si-yue WANG,Hui HUANG,Jin JIANG,Ying YE,Chun-lan HUANG,Yun ZHOU,Wen-jing GAO,Can-qing YU,Jun LV,Xiao-ling WU,Xiao-ming HUANG,Wei-hua CAO,Yan-sheng YAN,Tao WU,Li-ming LI. Bivariate heritability estimation of resting heart rate and common chronic disease based on extended pedigrees [J]. Journal of Peking University (Health Sciences), 2020, 52(3): 432-437. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 285
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 954
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
Cited |
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| Shared | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Discussed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
||