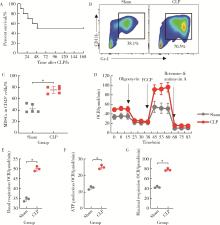
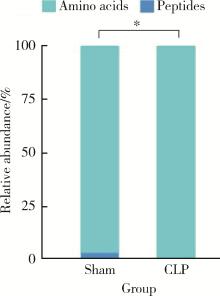
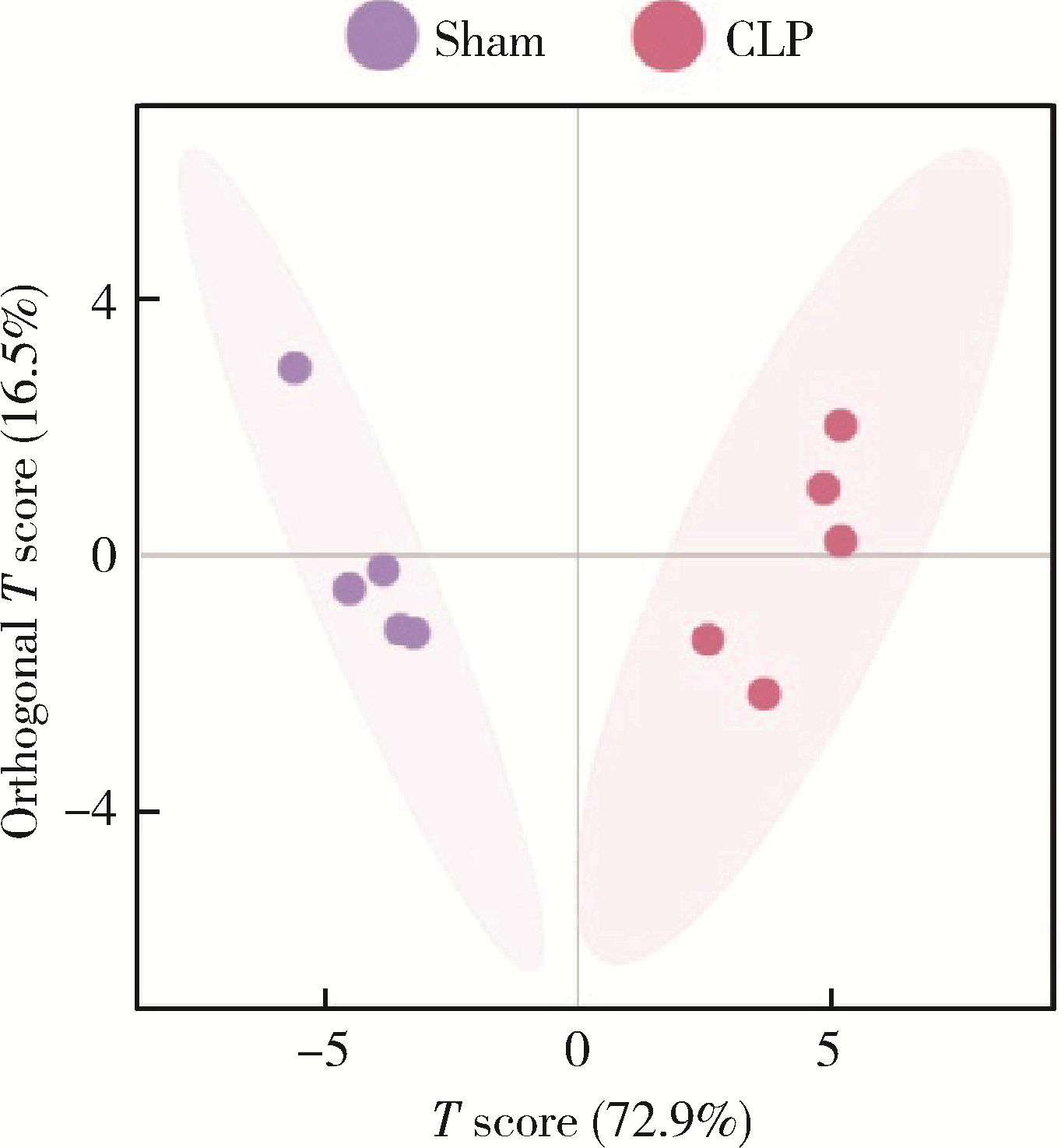
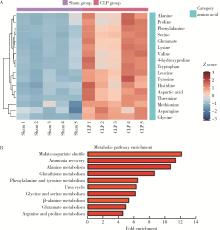
Journal of Peking University (Health Sciences) ›› 2022, Vol. 54 ›› Issue (3): 532-540. doi: 10.19723/j.issn.1671-167X.2022.03.020
Previous Articles Next Articles
Characteristics of amino acid metabolism in myeloid-derived suppressor cells in septic mice
Yuan MA1,Yue ZHANG2,3,Rui LI2,3,Shu-wei DENG2,3,Qiu-shi QIN1,Liu-luan ZHU1,2,3,*( )
)
- 1. Institute of Infectious Diseases, Peking University Ditan Teaching Hospital, Beijing 100015, China
2. Institute of Infectious Diseases, Beijing Ditan Hospital, Capital Medical University, Beijing 100015, China
3. Beijing Key Laboratory of Emerging Infectious Diseases, Beijing 100015, China
CLC Number:
- R363.1
| 1 |
Singer M , Deutschman CS , Seymour CW , et al. The third international consensus definitions for sepsis and septic shock (sepsis-3)[J]. JAMA, 2016, 315 (8): 801- 810.
doi: 10.1001/jama.2016.0287 |
| 2 |
徐静媛, 邱海波. 脓毒症治疗的现状与展望[J]. 国际流行病学传染病学杂志, 2021, 48 (4): 259- 262.
doi: 10.3760/cma.j.cn331340-20210618-00126 |
| 3 |
Rudd KE , Johnson SC , Agesa KM , et al. Global, regional, and national sepsis incidence and mortality, 1990—2017: analysis for the Global Burden of Disease Study[J]. Lancet, 2020, 395 (10219): 200- 211.
doi: 10.1016/S0140-6736(19)32989-7 |
| 4 |
Schrijver IT , Théroude C , Roger T . Myeloid-derived suppressor cells in sepsis[J]. Front Immunol, 2019, 10, 327.
doi: 10.3389/fimmu.2019.00327 |
| 5 |
Mira JC , Gentile LF , Mathias BJ , et al. Sepsis pathophysiology, chronic critical illness, and persistent inflammation-immuno-suppression and catabolism syndrome[J]. Crit Care Med, 2017, 45 (2): 253- 262.
doi: 10.1097/CCM.0000000000002074 |
| 6 |
Veglia F , Perego M , Gabrilovich D . Myeloid-derived suppressor cells coming of age[J]. Nat Immunol, 2018, 19 (2): 108- 119.
doi: 10.1038/s41590-017-0022-x |
| 7 |
Consonni FM , Porta C , Marino A , et al. Myeloid-Derived suppressor cells: ductile targets in disease[J]. Front Immunol, 2019, 10, 949.
doi: 10.3389/fimmu.2019.00949 |
| 8 |
Karin N . The Development and homing of myeloid-derived suppressor cells: from a two-stage model to a multistep narrative[J]. Front Immunol, 2020, 11, 557- 586.
doi: 10.3389/fimmu.2020.00557 |
| 9 |
Venet F , Monneret G . Advances in the understanding and treatment of sepsis-induced immunosuppression[J]. Nat Rev Nephrol, 2018, 14 (2): 121- 137.
doi: 10.1038/nrneph.2017.165 |
| 10 |
Mathias B , Delmas AL , Ozrazgat-Baslanti T , et al. Human myeloid-derived suppressor cells are associated with chronic immune suppression after severe sepsis/septic shock[J]. Ann Surg, 2017, 265 (4): 827- 834.
doi: 10.1097/SLA.0000000000001783 |
| 11 |
Darden DB , Bacher R , Brusko MA , et al. Single-cell RNA-seq of human myeloid-derived suppressor cells in late sepsis reveals multiple subsets with unique transcriptional responses: a pilot study[J]. Shock, 2021, 55 (5): 587- 595.
doi: 10.1097/SHK.0000000000001671 |
| 12 |
Wegiel B , Vuerich M , Daneshmandi S , et al. Metabolic switch in the tumor microenvironment determines immune responses to anti-cancer therapy[J]. Front Oncol, 2018, 8, 284.
doi: 10.3389/fonc.2018.00284 |
| 13 |
Won WJ , Deshane JS , Leavenworth JW , et al. Metabolic and functional reprogramming of myeloid-derived suppressor cells and their therapeutic control in glioblastoma[J]. Cell Stress, 2019, 3 (2): 47- 65.
doi: 10.15698/cst2019.02.176 |
| 14 |
Leinwand J , Miller G . Regulation and modulation of antitumor immunity in pancreatic cancer[J]. Nat Immunol, 2020, 21 (10): 1152- 1159.
doi: 10.1038/s41590-020-0761-y |
| 15 |
Grzywa TM , Sosnowska A , Matryba P , et al. Myeloid cell-derived arginase in cancer immune response[J]. Front Immunol, 2020, 11, 938.
doi: 10.3389/fimmu.2020.00938 |
| 16 |
Rittirsch D , Huber-Lang MS , Flierl MA , et al. Immunodesign of experimental sepsis by cecal ligation and puncture[J]. Nat Protoc, 2009, 4 (1): 31- 36.
doi: 10.1038/nprot.2008.214 |
| 17 |
Su L , Li H , Xie A , et al. Dynamic changes in amino acid concentration profiles in patients with sepsis[J]. PLoS One, 2015, 10 (4): e0121933.
doi: 10.1371/journal.pone.0121933 |
| 18 |
Gunst J , Vanhorebeek I , Thiessen SE , et al. Amino acid supplements in critically ill patients[J]. Pharmacol Res, 2018, 130, 127- 131.
doi: 10.1016/j.phrs.2017.12.007 |
| 19 |
Al-Khami AA , Rodriguez PC , Ochoa AC . Metabolic reprogramming of myeloid-derived suppressor cells (MDSCs) in cancer[J]. Oncoimmunology, 2016, 5 (8): e1200771.
doi: 10.1080/2162402X.2016.1200771 |
| 20 |
Mohammadpour H , MacDonald CR , McCarthy PL , et al. β2-adrenergic receptor signaling regulates metabolic pathways critical to myeloid-derived suppressor cell function within the TME[J]. Cell Rep, 2021, 37 (4): 109883.
doi: 10.1016/j.celrep.2021.109883 |
| 21 |
Altinok O , Poggio JL , Stein DE , et al. Malate-aspartate shuttle promotes l-lactate oxidation in mitochondria[J]. J Cell Physiol, 2020, 235 (3): 2569- 2581.
doi: 10.1002/jcp.29160 |
| 22 |
Mansouri S , Shahriari A , Kalantar H , et al. Role of malate dehydrogenase in facilitating lactate dehydrogenase to support the glycolysis pathway in tumors[J]. Biomed Rep, 2017, 6 (4): 463- 467.
doi: 10.3892/br.2017.873 |
| 23 |
Li X , Li Y , Yu Q , et al. Metabolic reprogramming of myeloid-derived suppressor cells: an innovative approach confronting challenges[J]. J Leukoc Biol, 2021, 110 (2): 257- 270.
doi: 10.1002/JLB.1MR0421-597RR |
| 24 |
Bozkus CC , Elzey BD , Crist SA , et al. Expression of cationic amino acid transporter 2 is required for myeloid-derived suppressor cell-mediated control of t cell immunity[J]. J Immunol, 2015, 195 (11): 5237- 5250.
doi: 10.4049/jimmunol.1500959 |
| 25 |
Srivastava MK , Sinha P , Clements VK , et al. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine[J]. Cancer Res, 2010, 70 (1): 68- 77.
doi: 10.1158/0008-5472.CAN-09-2587 |
| 26 |
Zhao H , Raines LN , Huang SC . Carbohydrate and amino acid metabolism as hallmarks for innate immune cell activation and function[J]. Cells, 2020, 9 (3): 562.
doi: 10.3390/cells9030562 |
| [1] | Yuanyuan ZENG,Yun XIE,Daonan CHEN,Ruilan WANG. Related factors of euthyroid sick syndrome in patients with sepsis [J]. Journal of Peking University (Health Sciences), 2024, 56(3): 526-532. |
| [2] | Zhi-wei LIU,Peng LIU,Fan-xing MENG,Tian-shui LI,Ying WANG,Jia-qi GAO,Zuo-yi ZHOU,Cong WANG,Bin ZHAO. Regulative effects of endogenous sulfur dioxide on oxidant stress in myocardium of rat with sepsis [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 582-586. |
| [3] | LIU Yu-qing, LU Jian, HAO Yi-chang, XIAO Chun-lei, MA Lu-lin. Predicting model based on risk factors for urosepsis after percutaneous nephrolithotomy [J]. Journal of Peking University(Health Sciences), 2018, 50(3): 507-513. |
| [4] | CHEN Wei, HU Fan-lei, LIU Hong-jiang, XU Li-ling, LI Ying-ni, LI Zhan-guo. Myeloid-derived suppressor cells promoted autologous B cell proliferation in rheumatoid arthritis [J]. Journal of Peking University(Health Sciences), 2017, 49(5): 819-823. |
| [5] | ZHANG Xin, RU Xi-fang, WANG Ying, LI Xing, SANG Tian, FENG Qi. Clinical characteristics of neonatal fungal sepsis in neonatal intensive care unit [J]. Journal of Peking University(Health Sciences), 2017, 49(5): 789-793. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 135
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 604
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
Cited |
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| Shared | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Discussed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
||