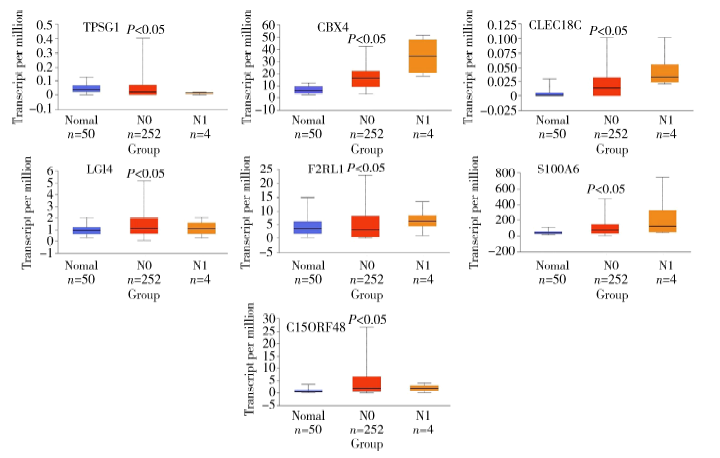
Journal of Peking University (Health Sciences) ›› 2020, Vol. 52 ›› Issue (2): 207-213. doi: 10.19723/j.issn.1671-167X.2020.02.003
Previous Articles Next Articles
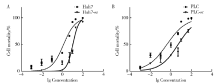
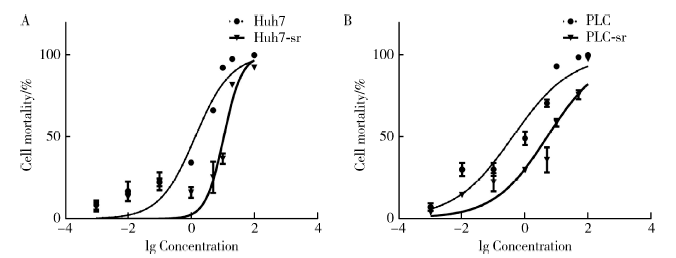
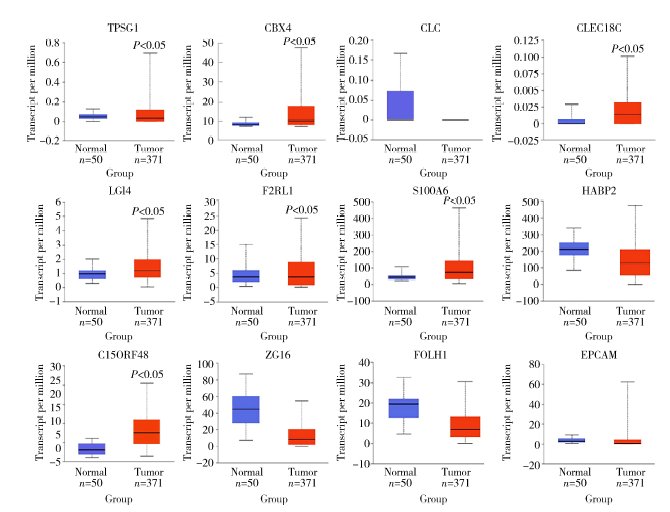
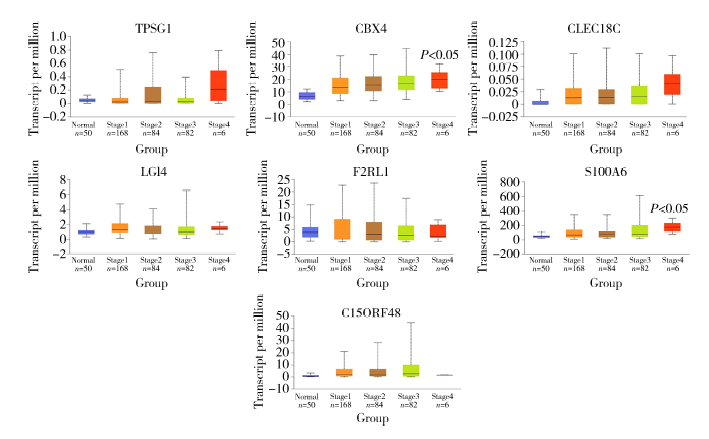
Establishment and gene expression analysis of drug-resistant cell lines in hepatocellular carcinoma induced by sorafenib
Bo MA1,Zhi-hua TIAN2,△( ),Li QU1,Yue-xiang LIU1,Hong ZHANG2,Hui-rong DING2
),Li QU1,Yue-xiang LIU1,Hong ZHANG2,Hui-rong DING2
- 1. Department of Lymphoma
2. Department of Laboratory, Ministry of Education Key Laboratory of Carcinogenesis and Translational Research,Peking University Cancer Hospital and Institute, Beijing 100142, China
CLC Number:
- R735.7
| [1] | Bray F, Ferlay J, Soerjomataram I , et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018,68(6):394-424. |
| [2] | Kulik L, El-Serag HB . Epidemiology and management of hepatocellular carcinoma[J]. Gastroenterology, 2019,156(2):477-491. |
| [3] | Llovet JM ,Zucman-rossi J,Pikarsky E, et al.Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2016,2(4):16018. |
| [4] | Wilhelm S, Carter C, Lynch M , et al. Discovery and development of sorafenib: a multik inase inhibitor for treating cancer[J]. Nat Reviews Drug Dis, 2006,5(10):835-844. |
| [5] | Keating GM, Santoro A . Sorafenib: a review of its use in advanced hepatocellular carcinoma[J]. Drugs, 2009,69(2):223-240. |
| [6] | Cheng AL, Kang YK, Chen Z , et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase Ⅲ randomised, double-blind, placebo-controlled trial[J]. Lancet Oncol, 2009,10(1):25-34. |
| [7] | Llovet JM, Ricci S, Mazzaferro V , et al. Sorafenib in advanced hepatocellular carcinoma[J]. N Engl J Med, 2008,359(4):378-390. |
| [8] | Zhang HM, Yang B, Guo B , et al. The role and practice of clinical bioinformatics in medical education[J]. Transpl Med J, 2018,3(4):175-178. |
| [9] | Darshan SC, Bhuwan B, Sai AHB , et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses[J]. Neoplasia, 2017,19:649-658. |
| [10] | Peng L, Bian XW, Li DK , et al. Large-scale RNA-seq transcriptome analysis of 4043 Cancers and 548 Normal tissue controls across 12 TCGA cancer types[J]. Sci Rep, 2015,5:13413. |
| [11] | Gebhardt ML, Mer AS , Andrade-Navarro MA. mBISON: finding miRNA target over-representation in gene lists from ChIP-sequencing data[J]. BMC Res Notes, 2015,16(8):157. |
| [12] | Kagey MH, Melhuish TA, Wotton D . The polycomb protein Pc2 is a SUMO E3[J]. Cell, 2003,113:127-137. |
| [13] | Li J, Xu Y, Long XD , et al. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity[J]. Cancer Cell, 2014,25:118-131. |
| [14] | Wang B, Tang J, Liao D , et al. Chromobox homolog 4 is corre-lated with prognosis and tumor cell growth in hepatocellular carcinoma[J]. Ann Surg Oncol, 2013,20(Suppl 3):S684-692. |
| [15] | Yang JL, Cheng DD, Zhu B , et al. Chromobox homolog 4 is positively correlated to tumor growth,survival and activation of HIF-1α signaling in human osteosarcoma under normoxic condition[J]. J Cancer, 2016,7(4):427-435. |
| [16] | Jiao HK, Xu Y, Li J , et al. Prognostic significance of Cbx4 expression and its beneficial effect for transarterial chemoembolization in hepatocellular carcinoma[J]. Cell Death Dis, 2015,6:e1689. |
| [1] | PANG Yong,ZHANG Sha,YANG Hua,ZHOU Rou-li. Serum LAPTM4B-35 protein as a novel diagnostic marker for hepatocellular carcinoma [J]. Journal of Peking University (Health Sciences), 2021, 53(4): 710-715. |
| [2] | Yun-bo XIE,Ji-yuan ZHANG,Mei-ling DU,Fan-ping MENG,Jun-liang FU,Li-min LIU,Song-shan WANG,Rui QU,Fang LIAN,Fei QIAO,Yang-liu CHEN,Ying-ying GAO,Ruo-nan XU,Ming SHI,Fu-sheng WANG. Efficacy and peripheral immunity analysis of allogeneic natural killer cells therapy in patients with hepatocellular carcinoma [J]. Journal of Peking University(Health Sciences), 2019, 51(3): 591-595. |
| [3] | Jing ZHANG,Jie CHEN,Gui-wen GUAN,Ting ZHANG,Feng-min LU,Xiang-mei CHEN. Expression and clinical significance of chemokine CXCL10 and its receptor CXCR3 in hepatocellular carcinoma [J]. Journal of Peking University(Health Sciences), 2019, 51(3): 402-408. |
|
||