Journal of Peking University (Health Sciences) ›› 2025, Vol. 57 ›› Issue (2): 245-252. doi: 10.19723/j.issn.1671-167X.2025.02.004
Previous Articles Next Articles
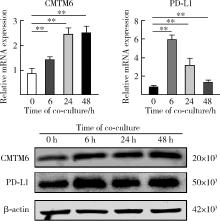
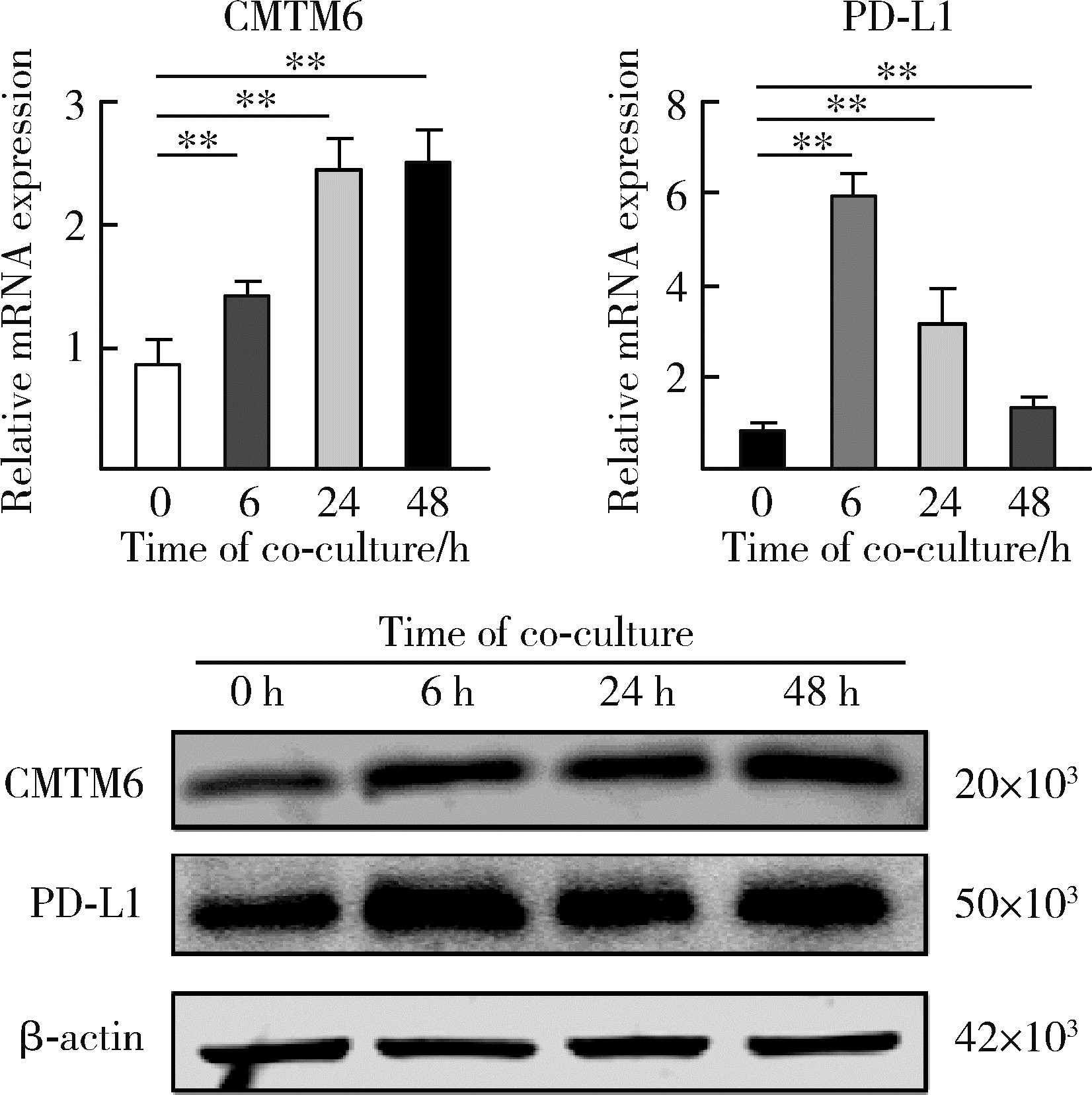
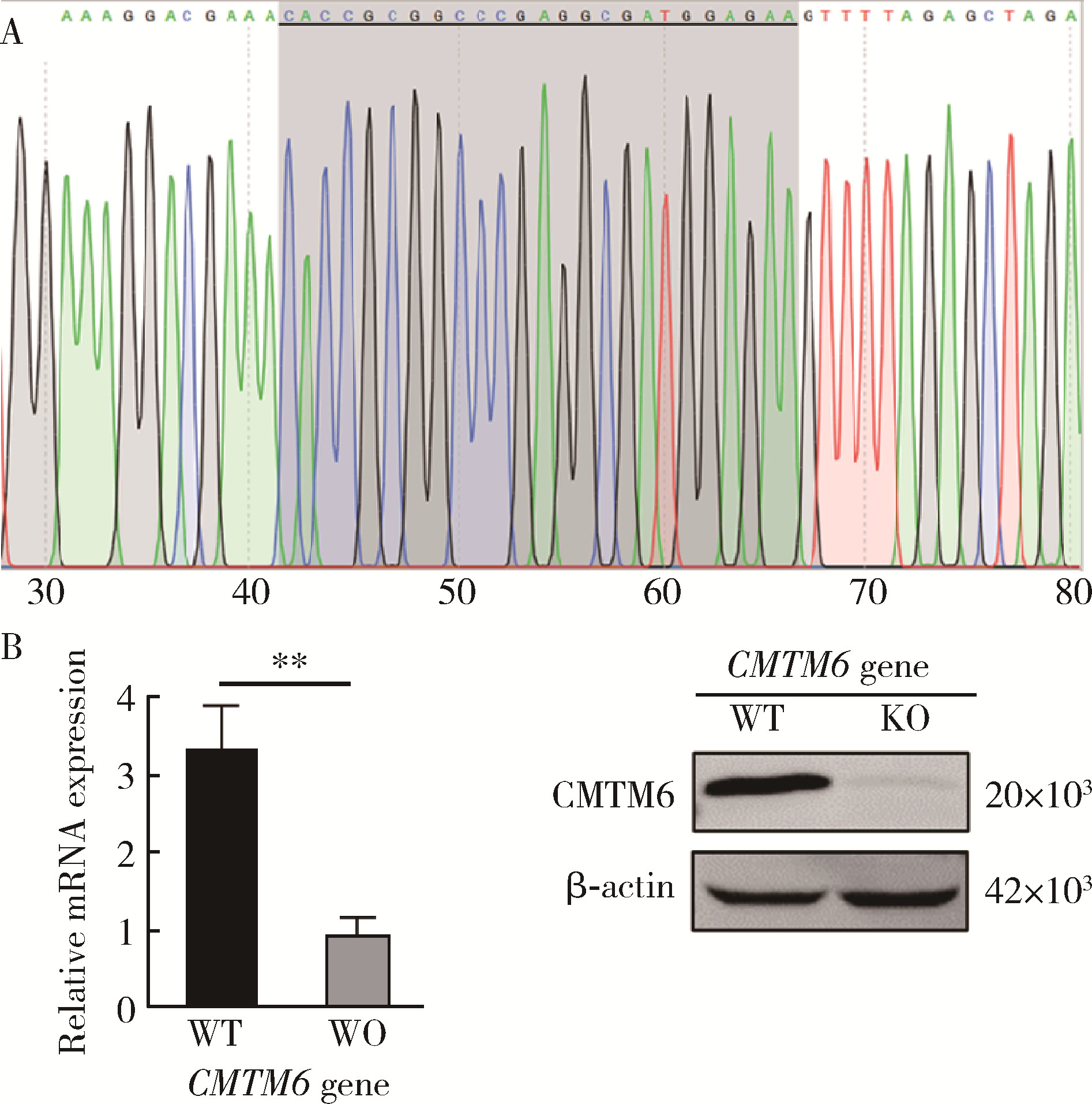
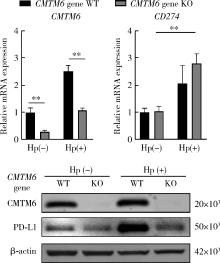
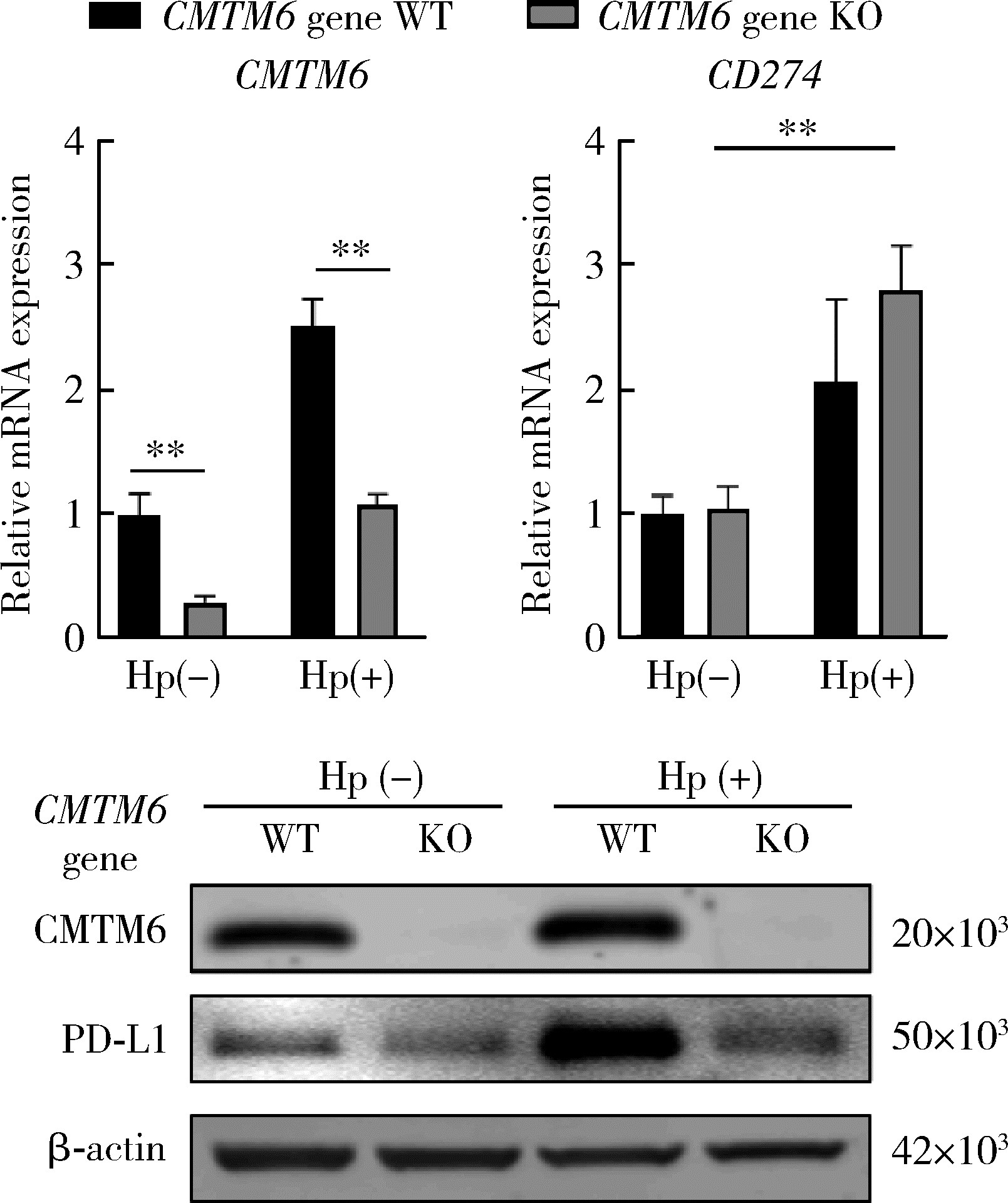
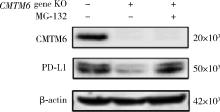
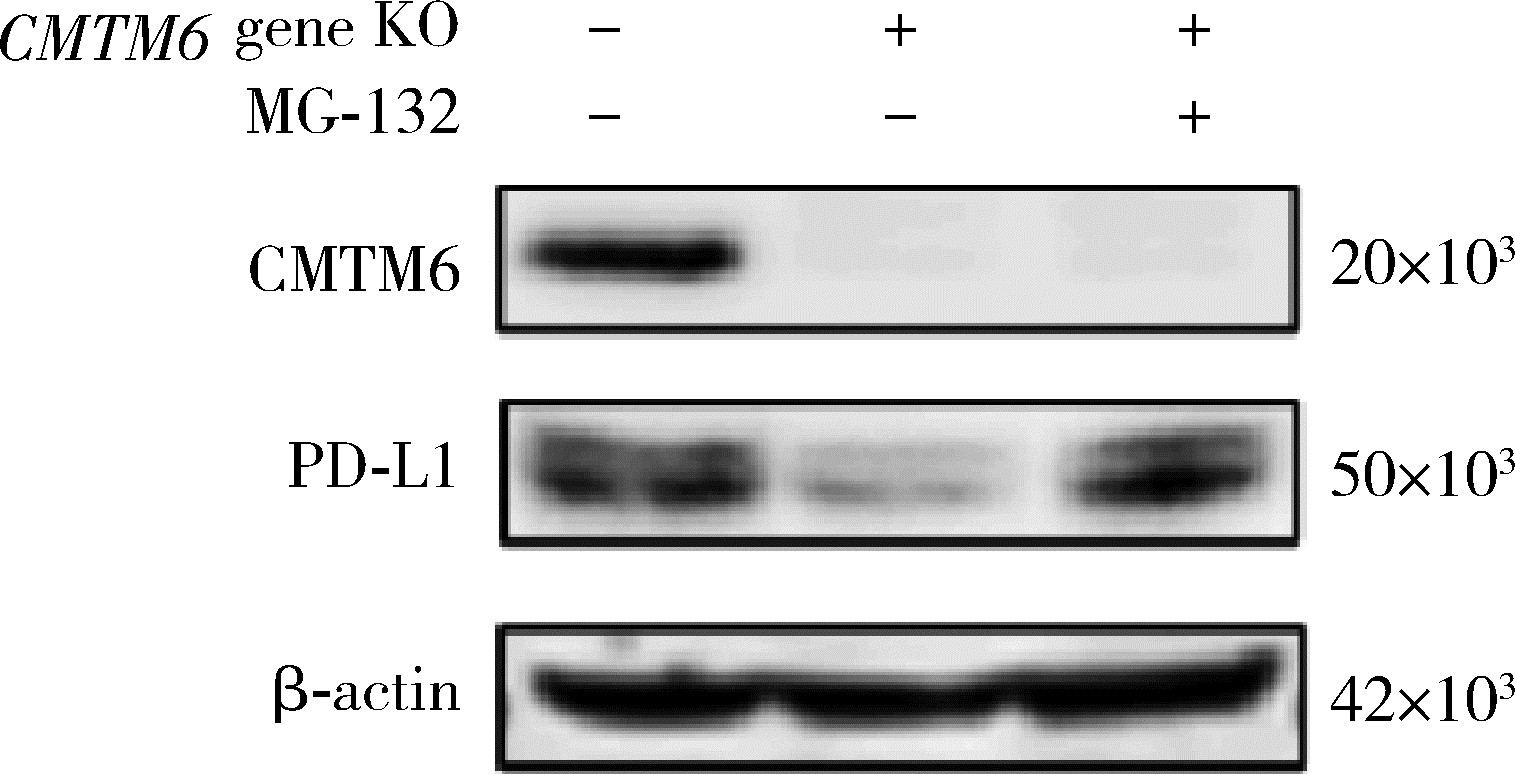
Effect of CMTM6 on PD-L1 in Helicobacter pylori infected gastric epithelial cells
Wei FU, Jing NING, Weiwei FU, Jing ZHANG*( ), Shigang DING*(
), Shigang DING*( )
)
- Department of Gastroenterology, Peking University Third Hospital; Beijing Key Laboratory for Helicobacter Pylori Infection and Upper Gastrointestinal Diseases, Beijing 100191, China
CLC Number:
- 735.2
| 1 |
Oster P , Vaillant L , Riva E , et al. Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies[J]. Gut, 2022, 71 (3): 457- 466.
doi: 10.1136/gutjnl-2020-323392 |
| 2 | Correa P . Human gastric carcinogenesis: A multistep and multifactorial process. First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention[J]. Cancer Res, 1992, 52 (24): 6735- 6740. |
| 3 |
Asaka M , Sugiyama T , Nobuta A , et al. Atrophic gastritis and intestinal metaplasia in Japan: Results of a large multicenter study[J]. Helicobacter, 2001, 6 (4): 294- 299.
doi: 10.1046/j.1523-5378.2001.00042.x |
| 4 | Zhao M , Liu Q , Liu W , et al. MicroRNA140 suppresses Helicobacter pyloripositive gastric cancer growth by enhancing the antitumor immune response[J]. Mol Med Rep, 2019, 20 (3): 2484- 2492. |
| 5 |
Xie G , Li W , Li R , et al. Helicobacter pylori promote B7-H1 expression by suppressing miR-152 and miR-200b in gastric cancer cells[J]. PLoS One, 2017, 12 (1): e0168822.
doi: 10.1371/journal.pone.0168822 |
| 6 |
Holokai L , Chakrabarti J , Broda T , et al. Increased programmed death-ligand 1 is an early epithelial cell response to Helicobacter pylori infection[J]. PLoS Pathog, 2019, 15 (1): e1007468.
doi: 10.1371/journal.ppat.1007468 |
| 7 |
Beswick EJ , Pinchuk IV , Das S , et al. Expression of the programmed death ligand 1, B7-H1, on gastric epithelial cells after Helicobacter pylori exposure promotes development of CD4+ CD25+ FoxP3+ regulatory T cells[J]. Infect Immun, 2007, 75 (9): 4334- 4341.
doi: 10.1128/IAI.00553-07 |
| 8 |
Wu YY , Lin CW , Cheng KS , et al. Increased programmed death-ligand-1 expression in human gastric epithelial cells in Helicobacter pylori infection[J]. Clin Exp Immunol, 2010, 161 (3): 551- 559.
doi: 10.1111/j.1365-2249.2010.04217.x |
| 9 |
Burr ML , Sparbier CE , Chan YC , et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity[J]. Nature, 2017, 549 (7670): 101- 105.
doi: 10.1038/nature23643 |
| 10 |
Mezzadra R , Sun C , Jae LT , et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators[J]. Nature, 2017, 549 (7670): 106- 110.
doi: 10.1038/nature23669 |
| 11 |
Guan X , Zhang C , Zhao J , et al. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas[J]. EBioMedicine, 2018, 35, 233- 243.
doi: 10.1016/j.ebiom.2018.08.012 |
| 12 |
Tian Y , Sun X , Cheng G , et al. The association of CMTM6 expression with prognosis and PD-L1 expression in triple-negative breast cancer[J]. Ann Transl Med, 2021, 9 (2): 131.
doi: 10.21037/atm-20-7616 |
| 13 |
Ishihara S , Iwasaki T , Kohashi K , et al. The association between the expression of PD-L1 and CMTM6 in undifferentiated pleomorphic sarcoma[J]. J Cancer Res Clin Oncol, 2021, 147 (7): 2003- 2011.
doi: 10.1007/s00432-021-03616-4 |
| 14 | Li X , Chen L , Gu C , et al. CMTM6 significantly relates to PD-L1 and predicts the prognosis of gastric cancer patients[J]. PeerJ, 2020 (8): e9536. |
| 15 |
Wang Z , Peng Z , Liu Q , et al. Co-expression with membrane CMTM6/4 on tumor epithelium enhances the prediction value of PD-L1 on anti-PD-1/L1 therapeutic efficacy in gastric adenocarcinoma[J]. Cancers (Basel), 2021, 13 (20): 5175.
doi: 10.3390/cancers13205175 |
| 16 |
Chen L , Yang QC , Li YC , et al. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma[J]. Cancer Immunol Res, 2020, 8 (2): 179- 191.
doi: 10.1158/2326-6066.CIR-19-0394 |
| 17 |
Huang X , Xiang L , Wang B , et al. CMTM6 promotes migration, invasion, and EMT by interacting with and stabilizing vimentin in hepatocellular carcinoma cells[J]. J Transl Med, 2021, 19 (1): 120.
doi: 10.1186/s12967-021-02787-5 |
| 18 |
Jin MH , Nam AR , Park JE , et al. Therapeutic co-targeting of WEE1 and ATM downregulates PD-L1 expression in pancreatic cancer[J]. Cancer Res Treat, 2020, 52 (1): 149- 166.
doi: 10.4143/crt.2019.183 |
| 19 | Tanaka E , Miyakawa Y , Kishikawa T , et al. Expression of circular RNA CDR1AS in colon cancer cells increases cell surface PDL1 protein levels[J]. Oncol Rep, 2019, 42 (4): 1459- 1466. |
| 20 |
Yamamoto Y , Kakizaki M , Shimizu T , et al. PD-L1 is induced on the hepatocyte surface via CKLF-like MARVEL transmembrane domain-containing protein 6 up-regulation by the anti-HBV drug entecavir[J]. Int Immunol, 2020, 32 (8): 519- 531.
doi: 10.1093/intimm/dxaa018 |
| 21 | Ho JY , Lu HY , Cheng HH , et al. UBE2S activates NF-κB signaling by binding with IκBα and promotes metastasis of lung adenocarcinoma cells[J]. Cell Oncol (Dordr), 2021, 44 (6): 1325- 1338. |
| [1] | Xue-li TIAN,Zhi-qiang SONG,Bao-jun SUO,Li-ya ZHOU,Cai-ling LI,Yu-xin ZHANG. Comparison of Epsilometer test and agar dilution method in detecting the sensitivity of Helicobacter pylori to metronidazole [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 934-938. |
| [2] | Wei-hua HOU,Shu-jie SONG,Zhong-yue SHI,Mu-lan JIN. Clinicopathological features of Helicobacter pylori-negative early gastric cancer [J]. Journal of Peking University (Health Sciences), 2023, 55(2): 292-298. |
| [3] | WANG Zi-jing,LI Zai-ling. Characteristics of gastric microbiota in children with Helicobacter pylori infection family history [J]. Journal of Peking University (Health Sciences), 2021, 53(6): 1115-1121. |
| [4] | YANG Guang, CHENG Qing-li, LI Chun-lin, JIA Ya-li, YUE Wen, PEI Xue-tao, LIU Yang, ZHAO Jia-hui, DU Jing, AO Qiang-guo. High glucose reduced the repair function of kidney stem cells conditional medium to the hypoxia-injured renal tubular epithelium cells [J]. Journal of Peking University(Health Sciences), 2017, 49(1): 125-130. |
| [5] | YU Jing-ting,MENG Huan-xin, LIU Kai-ning. An modified culture method of primary human gingival epithelial cells [J]. Journal of Peking University(Health Sciences), 2016, 48(4): 733-737. |
| [6] | REN Yu-Lan, ZHAN Yuan, LU Lu, LI Sheng-Lin, FU Xin, YU Guang-Yan, CAO Tong, LIU He. Expression characteristics of epithelial markers in human embryonic stem cells differentiating into keratinocytes [J]. Journal of Peking University(Health Sciences), 2015, 47(2): 305-311. |
| [7] | ZHONG Jin-Sheng, MEI Fang, ZHANG Hong-Quan. Comparison of biological characteristics of human gingival junctional epithelial cells and oral epithelial cells [J]. Journal of Peking University(Health Sciences), 2015, 47(1): 32-36. |
|
||