| [1] |
Kulkoyluoglu-Cotul E, Smith BP, Wrobel K , et al. Combined targeting of estrogen receptor Alpha and XPO1 prevent akt activation, remodel metabolic pathways and induce autophagy to overcome tamoxifen resistance[J]. Cancers (Basel), 2019,11(4):479.
|
| [2] |
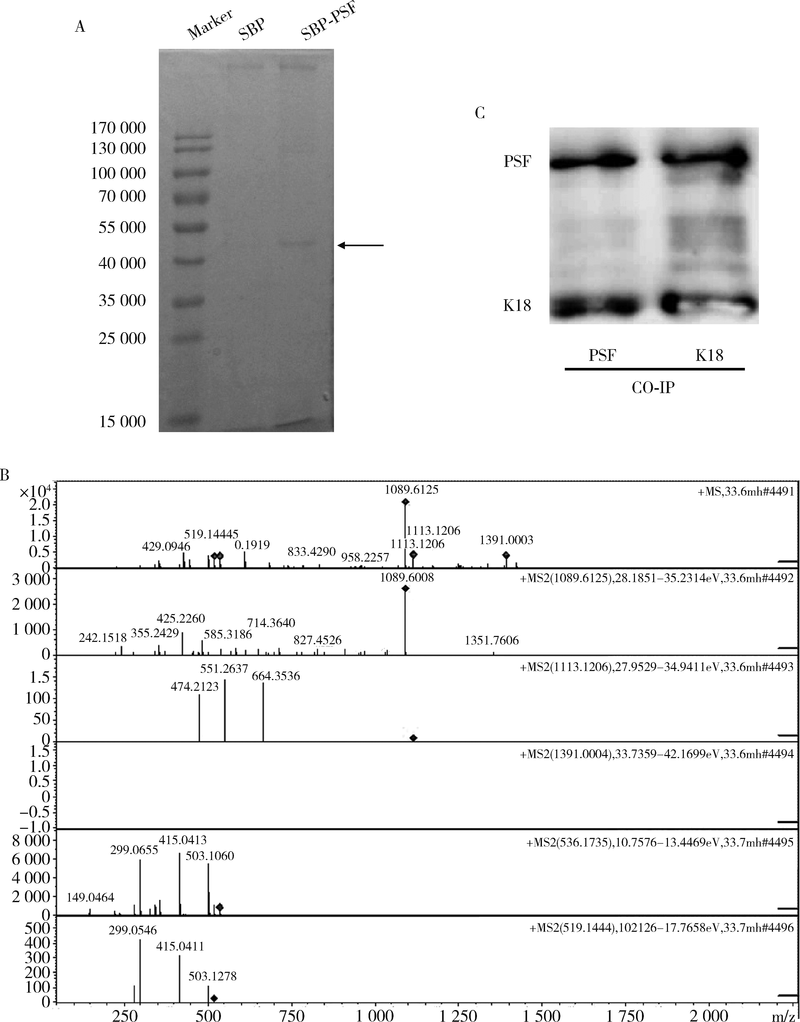
Ren S, She M, Li M , et al. The RNA/DNA-binding protein PSF relocates to cell membrane and contributes cells' sensitivity to antitumor drug, doxorubicin[J]. Cytometry A, 2014,85(3):231-241.
|
| [3] |
Knott GJ, Bond CS, Fox AH . The DBHS proteins SFPQ, NONO and PSPC1: A multipurpose molecular scaffold[J]. Nucleic Acids Res, 2016,44(9):3989-4004.
|
| [4] |
Gao Z, Chen M, Tian X , et al. A novel human lncRNA SANT1 cis-regulates the expression of SLC47A2 by altering SFPQ/E2F1/HDAC1 binding to the promoter region in renal cell carcinoma[J]. RNA Biol, 2019,16(7):940-949.
|
| [5] |
de Silva HC, Lin MZ, Phillips L , et al. IGFBP-3 interacts with NONO and SFPQ in PARP-dependent DNA damage repair in triple-negative breast cancer[J]. Cell Mol Life Sci, 2019,76(10):2015-2030.
|
| [6] |
Takayama K I, Suzuki T, Fujimura T , et al. Dysregulation of spliceosome gene expression in advanced prostate cancer by RNA-binding protein PSF[J]. Proc Natl Acad Sci USA, 2017,114(39):10461-10466.
|
| [7] |
Yarosh CA, Iacona JR, Lutz CS , et al. PSF: nuclear busy-body or nuclear facilitator?[J]. Wiley Interdiscip Rev RNA, 2015,6(4):351-367.
|
| [8] |
Izumi H, Mccloskey A, Shinmyozu K , et al. p54nrb/NonO and PSF promote U snRNA nuclear export by accelerating its export complex assembly[J]. Nucleic Acids Res, 2014,42(6):3998-4007.
|
| [9] |
Duan Y, Sun Y, Zhang F , et al. Keratin K18 increases cystic fibrosis transmembrane conductance regulator (CFTR) surface expression by binding to its C-terminal hydrophobic patch[J]. J Biol Chem, 2012,287(48):40547-40559.
|
| [10] |
Chen J, Ouyang H, An X , et al. Vault RNAs partially induces drug resistance of human tumor cells MCF-7 by binding to the RNA/DNA-binding protein PSF and inducing oncogene GAGE6[J]. PLoS One, 2018,13(1):e0191325.
|
| [11] |
Wang G, Cui Y, Zhang G , et al. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA[J]. Proc Natl Acad Sci USA, 2009,106(39):16794-16798.
|
| [12] |
Wang XT, Xia QY, Ni H , et al. SFPQ/PSF-TFE3 renal cell carcinoma: a clinicopathologic study emphasizing extended morphology and reviewing the differences between SFPQ-TFE3 RCC and the corresponding mesenchymal neoplasm despite an identical gene fusion[J]. Hum Pathol, 2017,63:190-200.
|
| [13] |
Ishikawa N, Nagase M, Takami S , et al. Xp11.2 translocation renal cell carcinoma with SFPQ/PSF-TFE3 fusion gene: A case report with unusual histopathologic findings[J]. Pathol Res Pract, 2019,215(9):152479.
|
| [14] |
Ha K, Takeda Y, Dynan WS . Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells[J]. DNA Repair (Amst), 2011,10(3):252-259.
|
| [15] |
Zou Y, He L, Wu CH , et al. PSF is an IbeA-binding protein contributing to meningitic Escherichia coli K1 invasion of human brain microvascular endothelial cells[J]. Med Microbiol Immunol, 2007,196(3):135-143.
|
| [16] |
Toivola DM, Boor P, Alam C , et al. Keratins in health and disease[J]. Curr Opin Cell Biol, 2015,32:73-81.
|
| [17] |
Ramot Y, Zlotogorski A . Keratins: the hair shaft's backbone revealed[J]. Exp Dermatol, 2015,24(6):416-417.
|
| [18] |
Lee S Y, Kim S, Lim Y , et al. Keratins regulate Hsp70-mediated nuclear localization of p38 mitogen-activated protein kinase[J/OL]. J Cell Sci, 2019, 9(2019-09-26)[2019-11-09].
|
| [19] |
Zhang B, Wang J, Liu W , et al. Cytokeratin 18 knockdown decreases cell migration and increases chemosensitivity in non-small cell lung cancer[J]. J Cancer Res Clin Oncol, 2016,142(12):2479-2487.
|
| [20] |
Shi R, Wang C, Fu N , et al. Downregulation of cytokeratin 18 enhances BCRP-mediated multidrug resistance through induction of epithelial-mesenchymal transition and predicts poor prognosis in breast cancer[J]. Oncol Rep, 2019,41(5):3015-3026.
|
 ),Lu-yao LONG1,2,Cheng-shan XU1
),Lu-yao LONG1,2,Cheng-shan XU1