Journal of Peking University (Health Sciences) ›› 2021, Vol. 53 ›› Issue (1): 24-33. doi: 10.19723/j.issn.1671-167X.2021.01.005
Previous Articles Next Articles
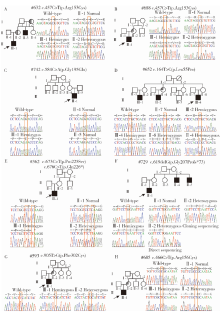
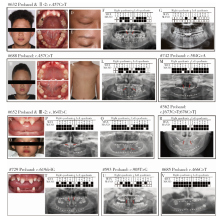
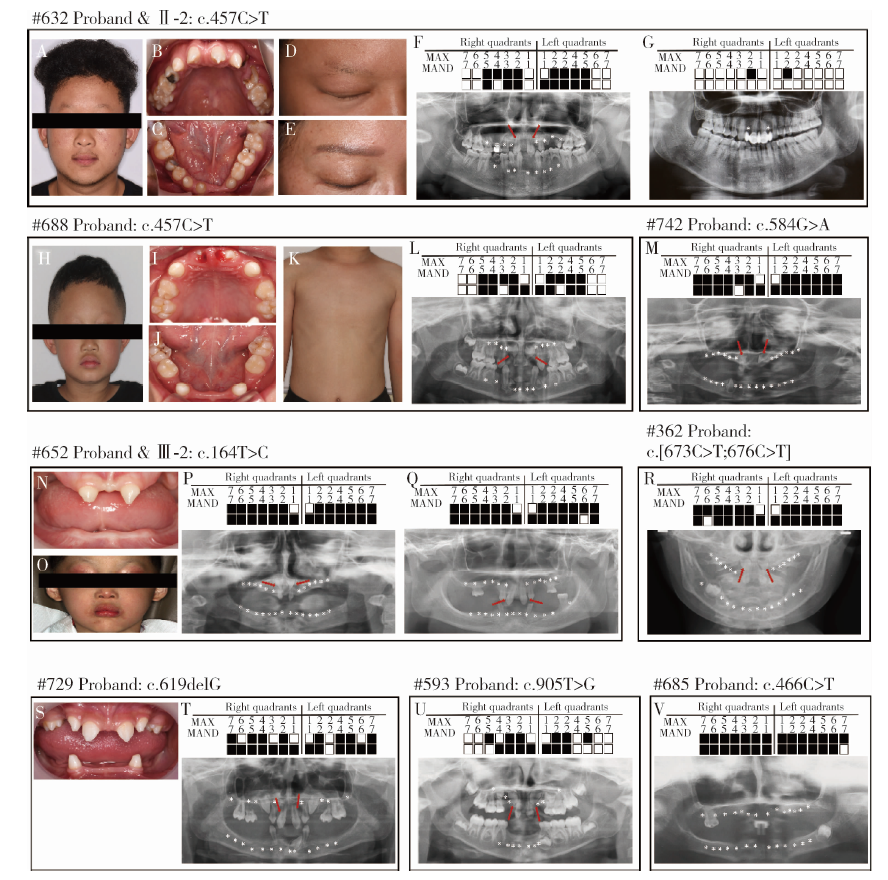
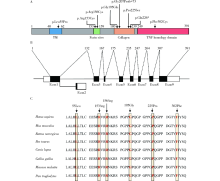
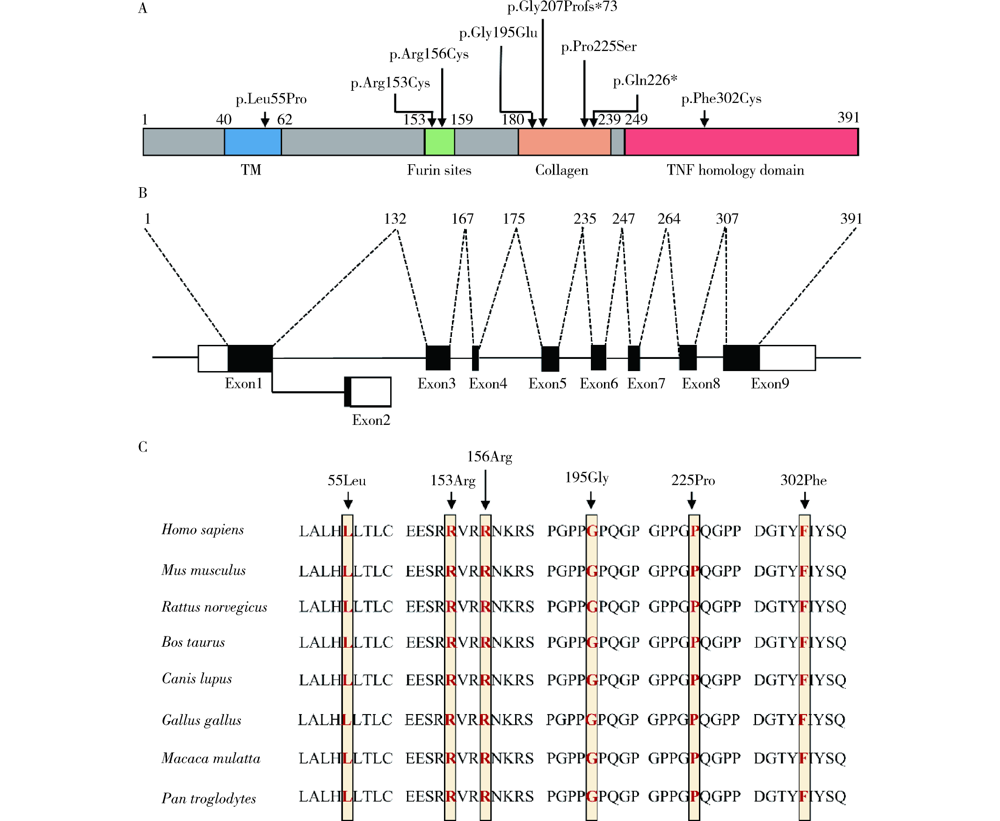
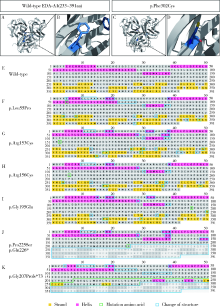
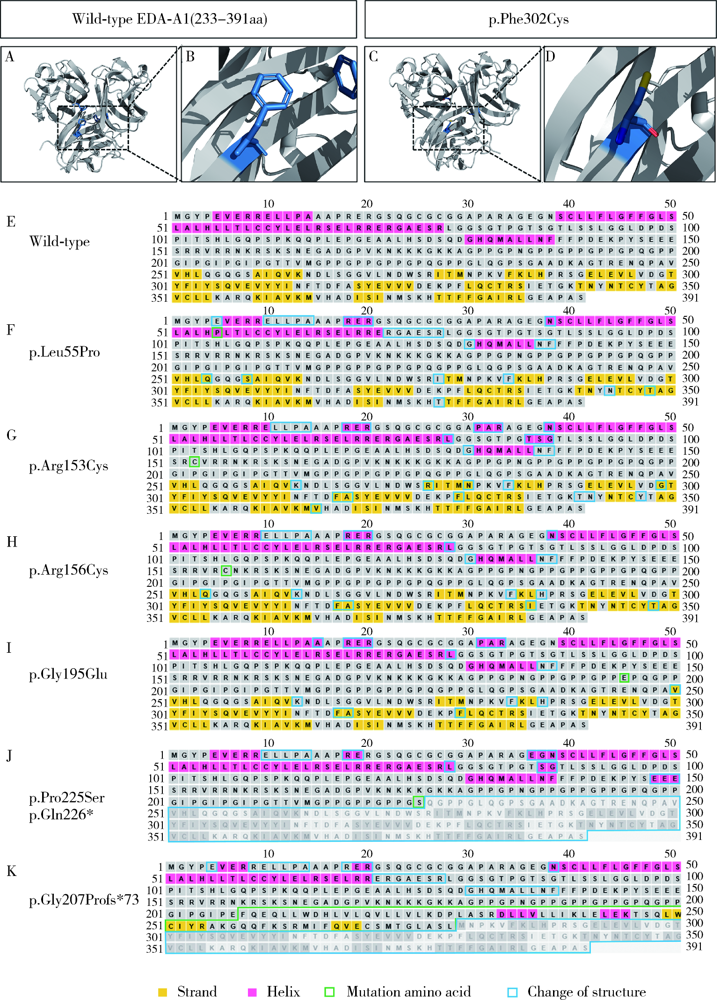
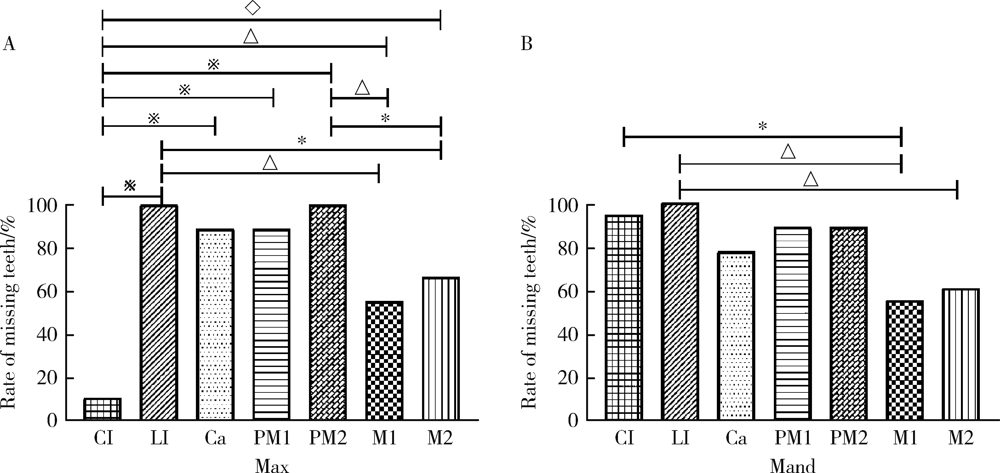
Detection of EDA gene mutation and phenotypic analysis in patients with hypohidrotic ectodermal dysplasia
WU Jun-yi1,YU Miao1,SUN Shi-chen1,2,FAN Zhuang-zhuang1,3,ZHENG Jing-lei1,ZHANG Liu-tao1,FENG Hai-lan1,LIU Yang1,Δ( ),HAN Dong1,Δ(
),HAN Dong1,Δ( )
)
- 1. Department of Prosthodontics, Peking University School and Hospital of Stomatology & National Clinical Research Center for Oral Disease & National Engineering Laboratory for Digital Material Technology of Stomatology & Beijing Key Laboratory of Digital Stomatology, Beijing 100081, China
2. Department of Dental Implantology & Prosthetic Dentistry, Shen-zhen Stomatology Hospital, Shenzhen 518001, Guangdong, China
3. Department of Stomatology, Beijing Hospital;National Center of Gerontology; Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, Beijing 100010, China
CLC Number:
- R394.1
| [1] |
Chandravanshi SL. Hypohidrotic ectodermal dysplasia: a case report[J]. Orbit, 2020,39(4):298-301.
doi: 10.1080/01676830.2019.1688358 pmid: 31694435 |
| [2] |
Anbouba GM, Carmany EP, Natoli JL. The characterization of hypodontia, hypohidrosis, and hypotrichosis associated with X-linked hypohidrotic ectodermal dysplasia: A systematic review[J]. Am J Med Genet A, 2020,182(4):831-841.
doi: 10.1002/ajmg.a.61493 pmid: 31981414 |
| [3] |
Noriega-Juárez MA, García-Delgado C, Villaseñor-Domínguez A, et al. X-linked hypohidrotic ectodermal dysplasia by a de novo recurrent variant in a Mexican patient[J]. Bol Med Hosp Infant Mex, 2020,77(4):212-217.
doi: 10.24875/BMHIM.19000209 pmid: 32713954 |
| [4] |
Abdulla AM, Almaliki AY, Shakeela NV, et al. Prosthodontic management of a pediatric patient with Christ-Siemens-Touraine Syndrome: a case report[J]. Int J Clin Pediatr Dent, 2019,12(6):569-572.
doi: 10.5005/jp-journals-10005-1697 pmid: 32440077 |
| [5] |
Martínez-Romero MC, Ballesta-Martínez MJ, López-González V, et al. EDA, EDAR, EDARADD and WNT10A allelic variants in patients with ectodermal derivative impairment in the Spanish population[J]. Orphanet J Rare Dis, 2019,14(1):281.
doi: 10.1186/s13023-019-1251-x pmid: 31796081 |
| [6] |
Park JS, Ko JM, Chae JH. Novel and private EDA mutations and clinical phenotypes of Korean patients with X-Linked hypohidrotic ectodermal dysplasia[J]. Cytogenet Genome Res, 2019,158(1):1-9.
doi: 10.1159/000500214 pmid: 31129666 |
| [7] |
Monreal AW, Ferguson BM, Headon DJ, et al. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia[J]. Nat Genet, 1999,22(4):366-369.
doi: 10.1038/11937 pmid: 10431241 |
| [8] |
Srivastava AK, Pispa J, Hartung AJ, et al. The tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains[J]. Proc Natl Acad Sci USA, 1997,94(24):13069-13074.
doi: 10.1073/pnas.94.24.13069 pmid: 9371801 |
| [9] |
Ezer S, Bayés M, Elomaa O, et al. Ectodysplasin is a collagenous trimeric type Ⅱ membrane protein with a tumor necrosis factor-like domain and co-localizes with cytoskeletal structures at lateral and apical surfaces of cells[J]. Hum Mol Genet, 1999,8(11):2079-2086.
doi: 10.1093/hmg/8.11.2079 pmid: 10484778 |
| [10] |
Schneider P, Street SL, Gaide O, et al. Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member ectodysplasin-A[J]. J Biol Chem, 2001,276(22):18819-18827.
doi: 10.1074/jbc.M101280200 pmid: 11279189 |
| [11] |
Huang SX, Liang JL, Sui WG, et al. EDA mutation as a cause of hypohidrotic ectodermal dysplasia: a case report and review of the literature[J]. Genet Mol Res, 2015,14(3):10344-10351.
doi: 10.4238/2015.August.28.21 pmid: 26345974 |
| [12] |
Li SM, Zhou J, Zhang LY, et al. Ectodysplasin A regulates epithelial barrier function through sonic hedgehog signalling pathway[J]. J Cell Mol Med, 2018,22(1):230-240.
doi: 10.1111/jcmm.13311 pmid: 28782908 |
| [13] |
Yamada A, Kawasaki M, Miake Y, et al. Overactivation of the NF-κB pathway impairs molar enamel formation[J]. Oral Dis, 2020,26(7):1513-1522.
doi: 10.1111/odi.13384 pmid: 32369672 |
| [14] |
Lévy J, Capri Y, Rachid M, et al. LEF1 haploinsufficiency causes ectodermal dysplasia[J]. Clin Genet, 2020,97(4):595-600.
doi: 10.1111/cge.13714 pmid: 32022899 |
| [15] |
Song SJ, Han D, Qu H, et al. EDA gene mutations underlie non-syndromic oligodontia[J]. J Dent Res, 2009,88(2):126-131.
doi: 10.1177/0022034508328627 pmid: 19278982 |
| [16] | Han Y, Wang XL, Zheng LY, et al. Pathogenic EDA mutations in Chinese Han families with hypohidrotic ectodermal dysplasia and genotype-phenotype: a correlation analysis[J]. Front Genet, 2020,11(21):1-11. |
| [17] |
Reyes-Reali J, Mendoza-Ramos MI, Garrido-Guerrero E, et al. Hypohidrotic ectodermal dysplasia: clinical and molecular review[J]. Int J Dermatol, 2018,57(8):965-972.
doi: 10.1111/ijd.14048 pmid: 29855039 |
| [18] |
Zhao J, Hua R, Zhao X, et al. Three novel mutations of the EDA gene in Chinese patients with X-linked hypohidrotic ectodermal dysplasia[J]. Br J Dermatol, 2008,158(3):614-617.
doi: 10.1111/j.1365-2133.2007.08383.x pmid: 18076698 |
| [19] |
Kowalczyk-Quintas C, Schneider P. Ectodysplasin A (EDA)-EDA receptor signalling and its pharmacological modulation[J]. Cytokine Growth Factor Rev, 2014,25(2):195-203.
doi: 10.1016/j.cytogfr.2014.01.004 pmid: 24508088 |
| [20] |
Savasta S, Carlone G, Castagnoli R, et al. X-Linked hypohidrotic ectodermal dysplasia: new features and a novel EDA gene mutation[J]. Cytogenet Genome Res, 2017,152(3):111-116.
doi: 10.1159/000478922 pmid: 28877528 |
| [21] |
Wahlbuhl M, Schuepbach-Mallepell S, Kowalczyk-Quintas C, et al. Attenuation of mammary gland dysplasia and feeding difficulties in tabby mice by fetal therapy[J]. J Mammary Gland Biol Neoplasia, 2018,23(3):125-138.
doi: 10.1007/s10911-018-9399-x pmid: 29855766 |
| [22] | Wahlbuhl-Becker M, Faschingbauer F, Beckmann MW, et al. Hypohidrotic ectodermal dysplasia: breastfeeding complications due to impaired breast development[J]. Geburtshilfe Frauen-heilkd, 2017,77(4):377-382. |
| [23] |
Wu CT, Morris JR. Genes, genetics, and epigenetics: a cor-respondence[J]. Science, 2001,293(5532):1103-1105.
doi: 10.1126/science.293.5532.1103 pmid: 11498582 |
| [24] |
Zhang J, Han D, Song SJ, et al. Correlation between the phenotypes and genotypes of X-linked hypohidrotic ectodermal dysplasia and non-syndromic hypodontia caused by ectodysplasin-A mutations[J]. Eur J Med Genet, 2011,54(4):e377-382.
doi: 10.1016/j.ejmg.2011.03.005 pmid: 21457804 |
| [25] |
Pispa J, Jung HS, Jernvall J, et al. Cusp patterning defect in tabby mouse teeth and its partial rescue by FGF[J]. Dev Biol, 1999,216(2):521-534.
pmid: 10642790 |
| [26] |
Cluzeau C, Hadj-Rabia S, Jambou M, et al. Only four genes (EDA1, EDAR, EDARADD, and WNT10A) account for 90% of hypohidrotic/anhidrotic ectodermal dysplasia cases[J]. Hum Mutat, 2011,32(1):70-77.
doi: 10.1002/humu.21384 pmid: 20979233 |
| [1] | Yun-fei SHI,Hao-jie WANG,Wei-ping LIU,Lan MI,Meng-ping LONG,Yan-fei LIU,Yu-mei LAI,Li-xin ZHOU,Xin-ting DIAO,Xiang-hong LI. Analysis of clinicopathological and molecular abnormalities of angioimmunoblastic T-cell lymphoma [J]. Journal of Peking University (Health Sciences), 2023, 55(3): 521-529. |
| [2] | Cai-peng QIN,Yu-xuan SONG,Meng-ting DING,Fei WANG,Jia-xing LIN,Wen-bo YANG,Yi-qing DU,Qing LI,Shi-jun LIU,Tao XU. Establishment of a mutation prediction model for evaluating the efficacy of immunotherapy in renal carcinoma [J]. Journal of Peking University (Health Sciences), 2022, 54(4): 663-668. |
| [3] | FENG Ke,NI Jing-jing,XIA Yan-qing,QU Xiao-wei,ZHANG Hui-juan,WAN Feng,HONG Kai,ZHANG Cui-lian,GUO Hai-bin. Genetic analysis of three cases of acephalic spermatozoa syndrome caused by SUN5 mutation and the outcome of assisted reproductive technology [J]. Journal of Peking University (Health Sciences), 2021, 53(4): 803-807. |
| [4] | Zhu YOU,Li-li XU,Xue-fen LI,Jian-yun ZHANG,Jing DU,Li-sha SUN. BRAF gene mutations in ameloblastic fibromas [J]. Journal of Peking University(Health Sciences), 2019, 51(1): 4-5. |
| [5] | Hao WANG,Yang LIU,Hao-chen LIU,Dong HAN,Hai-lan FENG. Detection and functional analysis of BMP2 gene mutation in patients with tooth agenesis [J]. Journal of Peking University(Health Sciences), 2019, 51(1): 9-15. |
|
||