Journal of Peking University(Health Sciences) ›› 2019, Vol. 51 ›› Issue (3): 556-563. doi: 10.19723/j.issn.1671-167X.2019.03.027
Previous Articles Next Articles
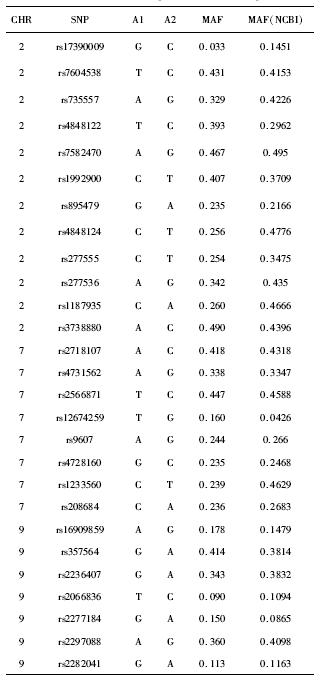
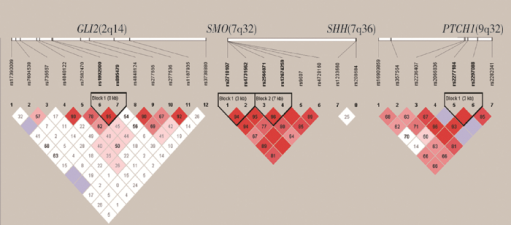
Analysis of single-nucleotide polymorphism of Sonic hedgehog signaling pathway in non-syndromic cleft lip and/or palate in the Chinese population
Jie-ni ZHANG1,*,Feng-qi SONG1,*,Shao-nan ZHOU1,Hui ZHENG1,Li-ying PENG1,Qian ZHANG2,Wang-hong ZHAO3,Tao-wen ZHANG4,Wei-ran LI1,Zhi-bo ZHOU5,Jiu-xiang LIN1△( ),Feng CHEN2△(
),Feng CHEN2△( )
)
- 1. Department of Orthodontics, Peking University School and Hospital of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Laboratory for Digital and Material Technology of Stomatology & Beijing Key Laboratory of Digital Stomatology, Beijing 100081, China
2. Department of Center Laboratory, Peking University School and Hospital of Stomatology, Beijing 100081, China
3. Department of Stomatology, Nanfang Hospital, Southern Medical University, Guangzhou 510000, China
4. Department of Orthodontics, Yantai Stomatological Hospital, Yantai 264000, Shandong, China
5. Department of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology, Beijing 100081, China
CLC Number:
- R393
| [1] |
Mossey PA, Little J, Munger RG , et al. Cleft lip and palate[J]. Lancet, 2009,374(9703):1773-1785.
doi: 10.1016/S0140-6736(09)60695-4 |
| [2] | Dixon MJ, Marazita ML, Beaty TH , et al. Cleft lip and palate: understanding genetic and environmental influences[J]. Nat Rev Genet, 2011,12(3):167-178. |
| [3] |
Lidral AC, Murray JC . Genetic approaches to identify disease genes for birth defects with cleft lip/palate as a model[J]. Birth Defects Res B, 2004,70(12):893-901.
doi: 10.1002/(ISSN)1542-0760 |
| [4] | Schliekelman P, Slatkin M . Multiplex relative risk and estimation of the number of loci underlying an inherited disease[J]. Am J Hum Genet, 2003,71(6):1369-1385. |
| [5] |
Ludwig KU, Mangold E, Herms S , et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci[J]. Nat Genet, 2012,44(9):968-971.
doi: 10.1038/ng.2360 |
| [6] |
Marazita ML, Mooney MP . Current concepts in the embryology and genetics of cleft lip and cleft palate[J]. Clin Plast Surg, 2004,31(2):125-140.
doi: 10.1016/S0094-1298(03)00138-X |
| [7] |
Zhang J, Zhou S, Zhang Q , et al. Proteomic analysis of RBP4/vitamin A in children with cleft lip and/or palate[J]. J Dent Res, 2014,93(6):547-552.
doi: 10.1177/0022034514530397 |
| [8] | Hu D, Helms JA . The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis[J]. Development, 1999,126(21):4873-4884. |
| [9] |
Vieira AR, Castilla EE, Ming JE , et al. Mutational analysis of the Sonic hedgehog gene in 220 newborns with oral clefts in a South American (ECLAMC) population[J]. Am J Med Genet, 2002,108(1):12-15.
doi: 10.1002/ajmg.10204 |
| [10] |
Han J, Mayo J, Xu X , et al. Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice[J]. Development, 2009,136(24):4225-4233.
doi: 10.1242/dev.036723 |
| [11] | Shimamura K, Hartigan DJ, Martinez S , et al. Longitudinal organization of the anterior neural plate and neural tube[J]. Development, 1995,121(12):3923-3933. |
| [12] |
Belloni E, Muenke M, Roessler E , et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly[J]. Nat Genet, 1996,14(3):353-356.
doi: 10.1038/ng1196-353 |
| [13] |
Chiang C, Litingtung Y, Lee E , et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function[J]. Nature, 1996,383(6599):407-413.
doi: 10.1038/383407a0 |
| [14] |
Rahnama F, Shimokawa T, Lauth M , et al. Inhibition of GLI1 gene activation by Patched1[J]. Biochem J, 2006,394(Pt 1):19-26.
doi: 10.1042/BJ20050941 |
| [15] |
Beaty TH, Fallin MD, Hetmanski JB , et al. Haplotype diversity in 11 candidate genes across four populations[J]. Genetics, 2005,171(1):259-267.
doi: 10.1534/genetics.105.043075 |
| [16] |
Xing J, Witherspoon DJ, Watkins WS , et al. HapMap tag SNP transferability in multiple populations: general guidelines[J]. Genomics, 2008,92(1):41-51.
doi: 10.1016/j.ygeno.2008.03.011 |
| [17] |
Jiang L, Zhang C, Li Y , et al. A non-synonymous polymorphism Thr115Met in the EpCAM gene is associated with an increased risk of breast cancer in Chinese population[J]. Breast Cancer Res Treat, 2011,126(2):487-495.
doi: 10.1007/s10549-010-1094-6 |
| [18] |
Ong KL, Li M, Tso AK , et al. Association of genetic variants in the adiponectin gene with adiponectin level and hypertension in Hong Kong Chinese[J]. Eur J Endocrinol, 2010,163(2):251-257.
doi: 10.1530/EJE-10-0251 |
| [19] |
Barrett JC, Fry B, Maller J , et al. Haploview: analysis and visualization of LD and haplotype maps[J]. Bioinformatics, 2005,21(2):263-265.
doi: 10.1093/bioinformatics/bth457 |
| [20] |
Farias LC, Gomes CC, Brito JA , et al. Loss of heterozygosity of the PTCH gene in ameloblastoma[J]. Hum Pathol, 2012,43(8):1229-1233.
doi: 10.1016/j.humpath.2011.08.026 |
| [21] |
Cohen MM . Holoprosencephaly: clinical, anatomic, and molecular dimensions.[J]. Birth Defects Res, 2006,76(9):658-673.
doi: 10.1002/(ISSN)1542-0760 |
| [22] | Carter TC, Molloy AM, Pangilinan F , et al. Testing reported associations of genetic risk factors for oral clefts in a large Irish study population[J]. Birth Defects Res, 2010,88(2):84-93. |
| [23] | Brand M, Heisenberg CP, Warga RM , et al. Mutations affecting development of the midline and general body shape during zebrafish embryogenesis[J]. Development, 1996,123(6):129-142. |
| [24] |
Eberhart JK, Swartz ME, Crump JG , et al. Early hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development[J]. Development, 2006,133(6):1069-1077.
doi: 10.1242/dev.02281 |
| [25] | Yahya MJ, Ismail PB, Nordin NB , et al. CNDP1, NOS3, and MnSOD polymorphisms as risk factors for diabetic nephropathy among type 2 diabetic patients in Malaysia[J]. J Nutr Metab, 2019(3):1-13. |
| [26] | Zhuo M, Zhuang X, Tang W , et al. Theimpact of IL-16 3’UTR polymorphism rs859 on lung carcinoma susceptibility among Chinese han individuals[J]. Biomed Res Int, 2018,12(24):1-10. |
| [27] |
Beaty TH, Hetmanski JB, Fallin MD , et al. Analysis of candidate genes on chromosome 2 in oral cleft case-parent trios from three populations[J]. Hum Genet, 2006,120(4):501-518.
doi: 10.1007/s00439-006-0235-9 |
| [28] |
Levi B, James AW, Nelson ER , et al. Role of Indian hedgehog signaling in palatal osteogenesis[J]. Plast Reconstr Surg, 2011,127(3):1182-1190.
doi: 10.1097/PRS.0b013e3182043a07 |
| [29] |
Helms JA, Kim CH, Hu D , et al. Sonic hedgehog participates in craniofacial morphogenesis and is down-regulated by teratogenic doses of retinoic acid[J]. Dev Biol, 1997,187(1):1-35.
doi: 10.1006/dbio.1997.8572 |
| [1] | Jing ZHANG,Jie CHEN,Gui-wen GUAN,Ting ZHANG,Feng-min LU,Xiang-mei CHEN. Expression and clinical significance of chemokine CXCL10 and its receptor CXCR3 in hepatocellular carcinoma [J]. Journal of Peking University(Health Sciences), 2019, 51(3): 402-408. |
| [2] | Zhi-ming SUN,Qian CHEN,Ming-hua LI,Wei-ning MA,Xu-yang ZHAO,Zhuo HUANG. Chronic phosphoproteomic in temporal lobe epilepsy mouse models induced by kainic acid [J]. Journal of Peking University(Health Sciences), 2019, 51(2): 197-205. |
| [3] | YANG Liu,CHU Xiao-yu,ZHAO Qi. Effects of RhoA on the adherens junction of murine ameloblasts [J]. Journal of Peking University(Health Sciences), 2018, 50(3): 521-526. |
| [4] | GONG Yan-qing, ZHANG Cui-jian, HE Shi-ming, LI Xue-song, ZhOU Li-qun, GUO Ying-lu. Nuclear export signal of androgen receptor regulated of androgen receptor stability in prostate cancer [J]. Journal of Peking University(Health Sciences), 2017, 49(4): 569-574. |
| [5] | CAI Yuan-fa, ZHANG Hua-ming, NIU Wen-yi, ZOU Yong-qiu, MA De-fu. Effects of equol on colon cancer cell proliferation [J]. Journal of Peking University(Health Sciences), 2017, 49(3): 383-387. |
|
||