Journal of Peking University(Health Sciences) ›› 2019, Vol. 51 ›› Issue (5): 805-812. doi: 10.19723/j.issn.1671-167X.2019.05.003
Previous Articles Next Articles
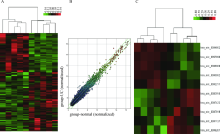
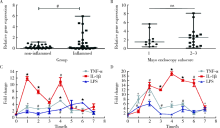
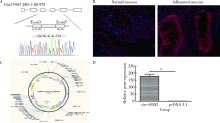
Effects of circular RNA circ-SOD2 on intestinal epithelial barrier and ulcerative colitis
Ting-ting WANG1,2,Ying HAN1,Fang-fang GAO1,Lei YE1,Yu-jun ZHANG1,△( )
)
- 1. The Central Laboratory, Peking University People’s Hospital, Beijing, 100044, China
2. Department of Gastroenterology, Peking University People’s Hospital, Beijing, 100044, China
CLC Number:
- R574.62
| [1] | Magro FGP, Eliakim R, Ardizzone S , et al. Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders[J]. J Crohns Colitis, 2017,11(6):649-670. |
| [2] | Ng SC, Shi HY, Hamidi N , et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies[J]. Lancet, 2017,390(10114):2769-2778. |
| [3] | Costello CM, Mah N, Hasler R , et al. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays[J]. PLoS Med, 2005,2(8):e199. |
| [4] | Ventham NT, Kennedy NA, Nimmo ER , et al. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics[J]. Gastroenterology, 2013,145(2):293-308. |
| [5] | Jeck WR, Sorrentino JA, Wang K , et al. Circular RNAs are abundant, conserved, and associated with ALU repeats[J]. RNA, 2013,19(2):141-157. |
| [6] | Salzman J, Chen RE, Olsen MN , et al. Cell-type specifc features of circular RNA expression[J]. PLoS Genet, 2013,9(9):e1003777. |
| [7] | Han D, Li J, Wang H , et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression[J]. Hepatology, 2017,66(4):1151-1164. |
| [8] | Guarnerio J, Bezzi M, Jeong JC , et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations[J]. Cell, 2016,165(2):289-302. |
| [9] | Liu Q, Zhang X, Hu X , et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation[J]. Sci Rep, 2016,6:22572. |
| [10] | Iparraguirre L, Munoz-Culla M, Prada-Luengo I , et al. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis[J]. Hum Mol Genet, 2017,26(18):3564-3572. |
| [11] | Qiao YQ, Cai CW, Shen J , et al. Circular RNA expression alterations in colon tissues of Crohn’s disease patients[J]. Mol Med Rep, 2019,19(5):4500-4506. |
| [12] | Yuan G, Chen T, Zhang H , et al. Comprehensive analysis of differential circular RNA expression in a mouse model of colitis-induced colon carcinoma[J]. Mol Carcinog, 2018,57(12):1825-1834. |
| [13] | Piwecka M, Glazar P, Hernandez-Miranda LR , et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function[J]. Science, 2017,357(6357):8526. |
| [14] | Min M, Peng L, Yang Y , et al. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a[J]. Inflamm Bowel Dis, 2014,20(4):652-659. |
| [15] | Wang H, Chao K, Ng SC , et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease[J]. Genome Biol, 2016,17:58. |
| [16] | He C, Yu T, Shi Y , et al. MicroRNA 301A promotes intestinal inflammation and colitis-associated cancer development by inhi-biting BTG1[J]. Gastroenterology, 2017,152(6):1434-1448. |
| [17] | Qu S, Yang X, Li X , et al. Circular RNA: a new star of non-coding RNAs[J]. Cancer Lett, 2015,365(2):141-148. |
| [18] | Glažar P, Papavasileiou P, Rajewsky N . circBase: a database for circular RNAs[J]. RNA, 2014,20(11):1666-1670. |
| [19] | Kramer MC, Liang D, Tatomer DC , et al. Combinatorial control of drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins[J]. Genes Dev, 2015,29(20):2168-2182. |
| [20] | Gerasimenko TN, Senyavina NV, Anisimov NU , et al. A model of cadmium uptake and transport in Caco-2 cells[J]. Bull Exp Biol Med, 2016,161(1):187-192. |
| [21] | Mirza AH, Berthelsen CH, Seemann SE , et al. Transcriptomic landscape of lncRNAs in inflammatory bowel disease[J]. Genome Med, 2015,7(1):39. |
| [22] | Wu F, Huang Y, Dong F , et al. Ulcerative colitis-associated long noncoding RNA, BC012900, regulates intestinal epithelial cell apoptosis[J]. Inflamm Bowel Dis, 2016,22(4):782-795. |
| [23] | Hammond SM . An overview of microRNAs[J]. Adv Drug Deliv Rev, 2015,87:3-14. |
| [1] | Bing-jie HE,Zhi-ke LIU,Peng SHEN,Ye-xiang SUN,Bin CHEN,Si-yan ZHAN,Hong-bo LIN. Epidemiological study on the incidence of inflammatory bowel disease in Yinzhou District, Ningbo City from 2011 to 2020 [J]. Journal of Peking University (Health Sciences), 2022, 54(3): 511-519. |
|
||