Journal of Peking University (Health Sciences) ›› 2020, Vol. 52 ›› Issue (5): 809-814. doi: 10.19723/j.issn.1671-167X.2020.05.003
Previous Articles Next Articles
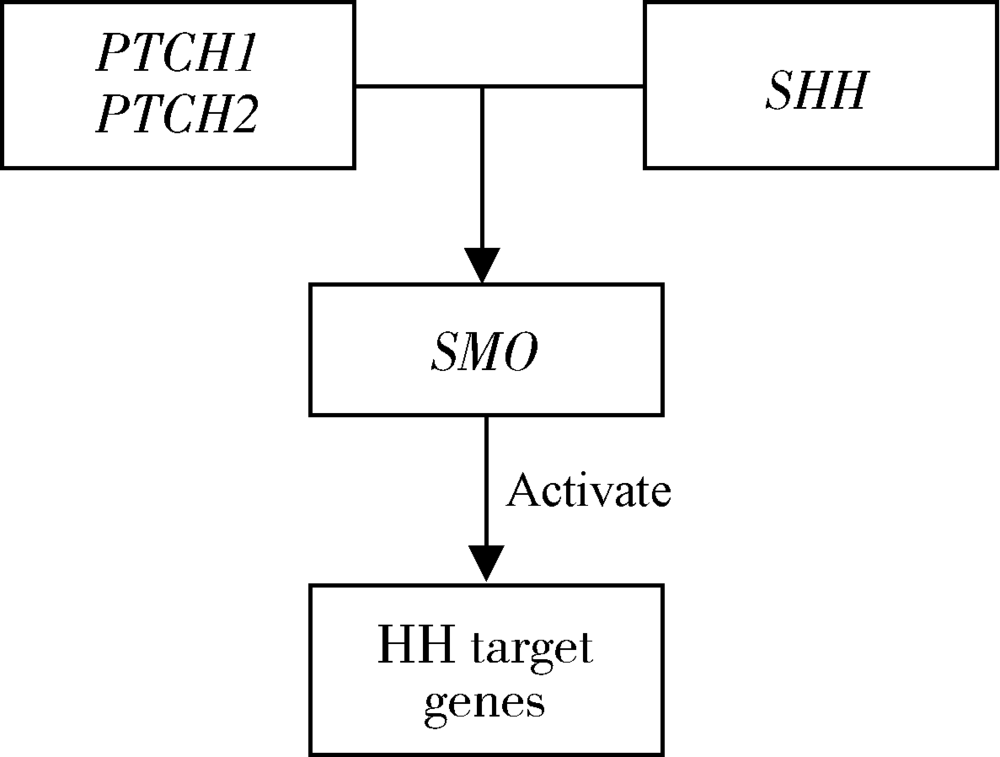
Exploring parent-of-origin effects for non-syndromic cleft lip with or without cleft palate on PTCH1, PTCH2, SHH, SMO genes in Chinese case-parent trios
Wen-yong LI1,Meng-ying WANG1,Ren ZHOU1,Si-yue WANG1,Hong-chen ZHENG1,Hong-ping ZHU2,Zhi-bo ZHOU2,Tao WU1,3,∆( ),Hong WANG1,Bing SHI4
),Hong WANG1,Bing SHI4
- 1. Department of Epidemiology and Biostatistics, School of Public Health, Peking University, Beijing 100191, China
2. Department of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Laboratory for Digital and Material Technology of Stomatology & Beijing Key Laboratory of Digital Stomatology, Beijing 100081, China
3. Key Laboratory of Reproductive Health, Ministry of Health, Beijing 100191, China
4. Department of Oral and Maxillofacial Surgery, State Key Laboratory of Oral Disease, West China College of Stomatology, Sichuan University, Chengdu 610041, China
CLC Number:
- R394
| [1] |
Wang M, Yuan Y, Wang Z, et al. Prevalence of orofacial clefts among live births in China: a systematic review and meta-analysis[J]. Birth Defects Res, 2017,109(13):1011-1019.
doi: 10.1002/bdr2.1043 pmid: 28635078 |
| [2] |
Harville EW, Wilcox AJ, Lie RT, et al. Cleft lip and palate versus cleft lip only: are they distinct defects[J]. Am J Epidemiol, 2005,162(5):448-453.
doi: 10.1093/aje/kwi214 pmid: 16076837 |
| [3] |
Leslie EJ, Marazita ML. Genetics of cleft lip and cleft palate[J]. Am J Med Genet C Semin Med Genet, 2013,163c(4):246-258.
doi: 10.1002/ajmg.c.31381 pmid: 24124047 |
| [4] |
Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms[J]. Dev Dyn, 2006,235(5):1152-1166.
doi: 10.1002/dvdy.20646 pmid: 16292776 |
| [5] |
Mossey PA, Little J, Munger RG, et al. Cleft lip and palate[J]. Lancet, 2009,374(9703):1773-1785.
doi: 10.1016/S0140-6736(09)60695-4 pmid: 19747722 |
| [6] |
Grant SF, Wang K, Zhang H, et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24[J]. J Pediatr, 2009,155(6):909-913.
doi: 10.1016/j.jpeds.2009.06.020 pmid: 19656524 |
| [7] |
Beaty TH, Marazita ML, Leslie EJ. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow's opportunities[J]. F1000Res, 2016,5:2800.
doi: 10.12688/f1000research.9503.1 pmid: 27990279 |
| [8] |
Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases[J]. Nature, 2009,461(7265):747-753.
doi: 10.1038/nature08494 pmid: 19812666 |
| [9] |
Guilmatre A, Sharp AJ. Parent of origin effects[J]. Clin Genet, 2012,81(3):201-209.
doi: 10.1111/j.1399-0004.2011.01790.x |
| [10] |
Yu Y, Zuo X, He M, et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity[J]. Nat Commun, 2017,8:14364.
doi: 10.1038/ncomms14364 pmid: 28232668 |
| [11] |
Beaty TH, Murray JC, Marazita ML, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4[J]. Nat Genet, 2010,42(6):525-529.
doi: 10.1038/ng.580 pmid: 20436469 |
| [12] |
Gjessing HK, Lie RT. Case-parent triads: estimating single- and double-dose effects of fetal and maternal disease gene haplotypes[J]. Ann Hum Genet, 2006,70(Pt 3):382-396.
doi: 10.1111/j.1529-8817.2005.00218.x pmid: 16674560 |
| [13] |
Gjerdevik M, Haaland OA, Romanowska J, et al. Parent-of-origin-environment interactions in case-parent triads with or without independent controls[J]. Ann Hum Genet, 2018,82(2):60-73.
doi: 10.1111/ahg.12224 pmid: 29094765 |
| [14] |
Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads[J]. Am J Hum Genet, 1999,65(1):229-235.
doi: 10.1086/302466 pmid: 10364536 |
| [15] |
Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease[J]. Nat Rev Mol Cell Biol, 2013,14(7):416-429.
doi: 10.1038/nrm3598 |
| [16] |
Taipale J, Cooper MK, Maiti T, et al. Patched acts catalytically to suppress the activity of Smoothened[J]. Nature, 2002,418(6900):892-897.
doi: 10.1038/nature00989 pmid: 12192414 |
| [17] |
Wantia N, Rettinger G. The current understanding of cleft lip malformations[J]. Facial Plast Surg, 2002,18(3):147-153.
doi: 10.1055/s-2002-33061 pmid: 12152133 |
| [18] |
Grosen D, Bille C, Petersen I, et al. Risk of oral clefts in twins[J]. Epidemiology, 2011,22(3):313-319.
doi: 10.1097/EDE.0b013e3182125f9c |
| [19] |
Mangold E, Ludwig KU, Birnbaum S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate[J]. Nat Genet, 2010,42(1):24-26.
doi: 10.1038/ng.506 pmid: 20023658 |
| [20] |
Sun Y, Huang Y, Yin A, et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate[J]. Nat Commun, 2015,6:6414.
pmid: 25775280 |
| [21] |
Lo Muzio L. Nevoid basal cell carcinoma syndrome (Gorlin syndrome)[J]. Orphanet J Rare Dis, 2008,3(1):32.
doi: 10.1186/1750-1172-3-32 |
| [22] |
Metzis V, Courtney AD, Kerr MC, et al. Patched1 is required in neural crest cells for the prevention of orofacial clefts[J]. Hum Mol Genet, 2013,22(24):5026-5035.
doi: 10.1093/hmg/ddt353 |
| [23] |
Xiao Y, Taub MA, Ruczinski I, et al. Evidence for SNP-SNP interaction identified through targeted sequencing of cleft case-parent trios[J]. Genet Epidemiol, 2017,41(3):244-250.
pmid: 28019042 |
| [24] |
de Araujo TK, Secolin R, Felix TM, et al. A multicentric association study between 39 genes and nonsyndromic cleft lip and palate in a Brazilian population[J]. J Craniomaxillofac Surg, 2016,44(1):16-20.
pmid: 26602496 |
| [25] |
Rubini M, Brusati R, Garattini G, et al. Cystathionine beta-synthase c.844ins68 gene variant and non-syndromic cleft lip and palate[J]. Am J Med Genet A, 2005,136a(4):368-372.
pmid: 16007597 |
| [26] |
Reutter H, Birnbaum S, Mende M, et al. TGFB3 displays parent-of-origin effects among central Europeans with nonsyndromic cleft lip and palate[J]. J Hum Genet, 2008,53(7):656-661.
doi: 10.1007/s10038-008-0296-9 |
| [27] |
Sull JW, Liang KY, Hetmanski JB, et al. Differential parental transmission of markers in RUNX2 among cleft case-parent trios from four populations[J]. Genet Epidemiol, 2008,32(6):505-512.
pmid: 18357615 |
| [28] |
Sull JW, Liang KY, Hetmanski JB, et al. Maternal transmission effects of the PAX genes among cleft case-parent trios from four populations[J]. Eur J Hum Genet, 2009,17(6):831-839.
pmid: 19142206 |
| [29] |
Sull JW, Liang KY, Hetmanski JB, et al. Evidence that TGFA influences risk to cleft lip with/without cleft palate through unconventional genetic mechanisms[J]. Hum Genet, 2009,126(3):385-394.
pmid: 19444471 |
| [30] |
Suazo J, Santos JL, Jara L, et al. Parent-of-origin effects for MSX1 in a Chilean population with nonsyndromic cleft lip/palate[J]. Am J Med Genet A, 2010,152a(8):2011-2016.
pmid: 20635363 |
| [31] |
Shi M, Murray JC, Marazita ML, et al. Genome wide study of maternal and parent-of-origin effects on the etiology of orofacial clefts[J]. Am J Med Genet A, 2012,158a(4):784-794.
doi: 10.1002/ajmg.a.35257 |
| [32] |
Garg P, Ludwig KU, Bohmer AC, et al. Genome-wide analysis of parent-of-origin effects in non-syndromic orofacial clefts[J]. Eur J Hum Genet, 2014,22(6):822-830.
doi: 10.1038/ejhg.2013.235 |
| [33] |
Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles[J]. Genet Epidemiol, 2002,23(3):221-233.
pmid: 12384975 |
| [1] | Enci XUE, Xi CHEN, Xueheng WANG, Siyue WANG, Mengying WANG, Jin LI, Xueying QIN, Yiqun WU, Nan LI, Jing LI, Zhibo ZHOU, Hongping ZHU, Tao WU, Dafang CHEN, Yonghua HU. Single nucleotide polymorphism heritability of non-syndromic cleft lip with or without cleft palate in Chinese population [J]. Journal of Peking University (Health Sciences), 2024, 56(5): 775-780. |
| [2] | Tianjiao HOU,Zhibo ZHOU,Zhuqing WANG,Mengying WANG,Siyue WANG,Hexiang PENG,Huangda GUO,Yixin LI,Hanyu ZHANG,Xueying QIN,Yiqun WU,Hongchen ZHENG,Jing LI,Tao WU,Hongping ZHU. Gene-gene/gene-environment interaction of transforming growth factor-β signaling pathway and the risk of non-syndromic oral clefts [J]. Journal of Peking University (Health Sciences), 2024, 56(3): 384-389. |
|
||