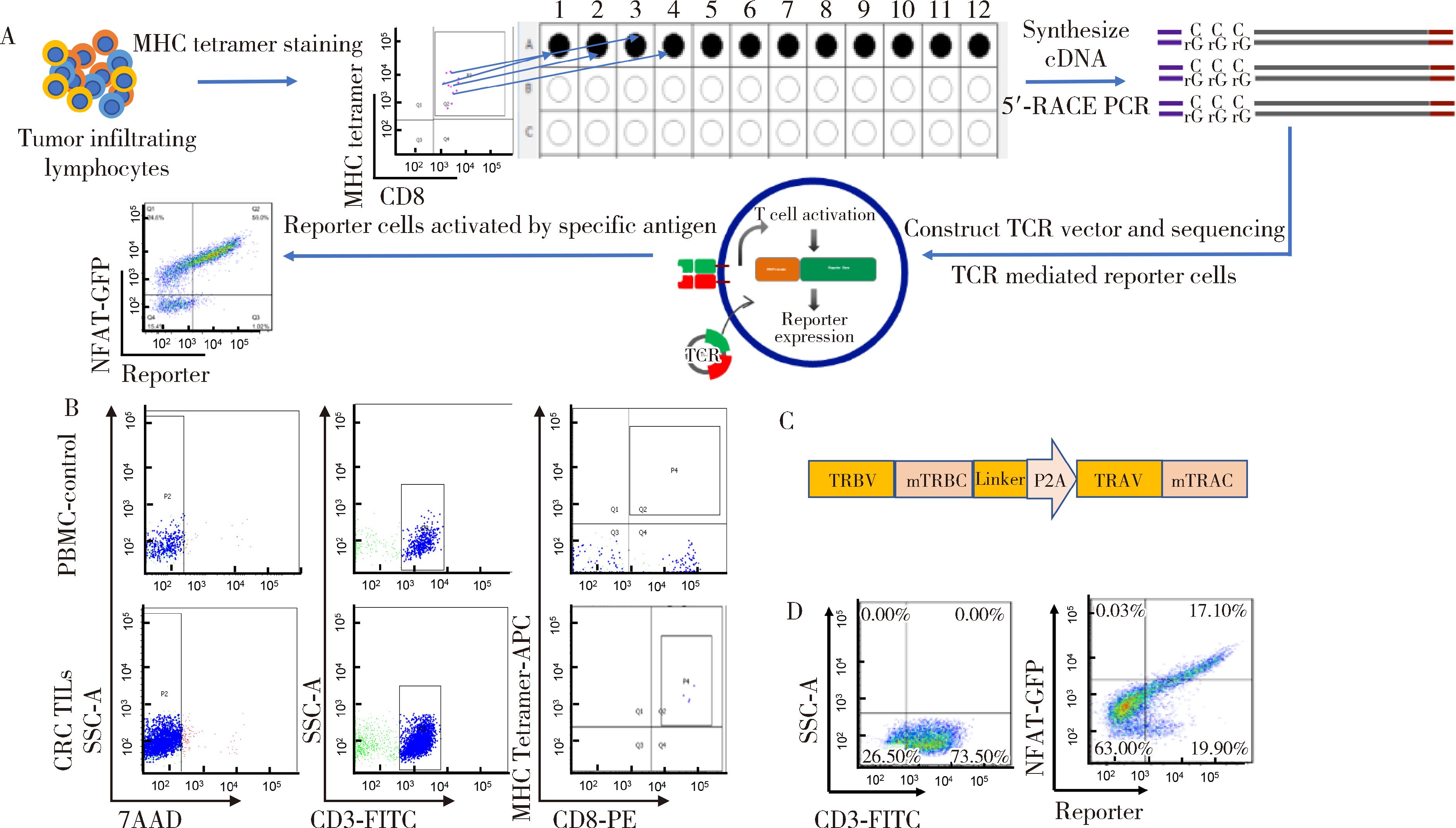
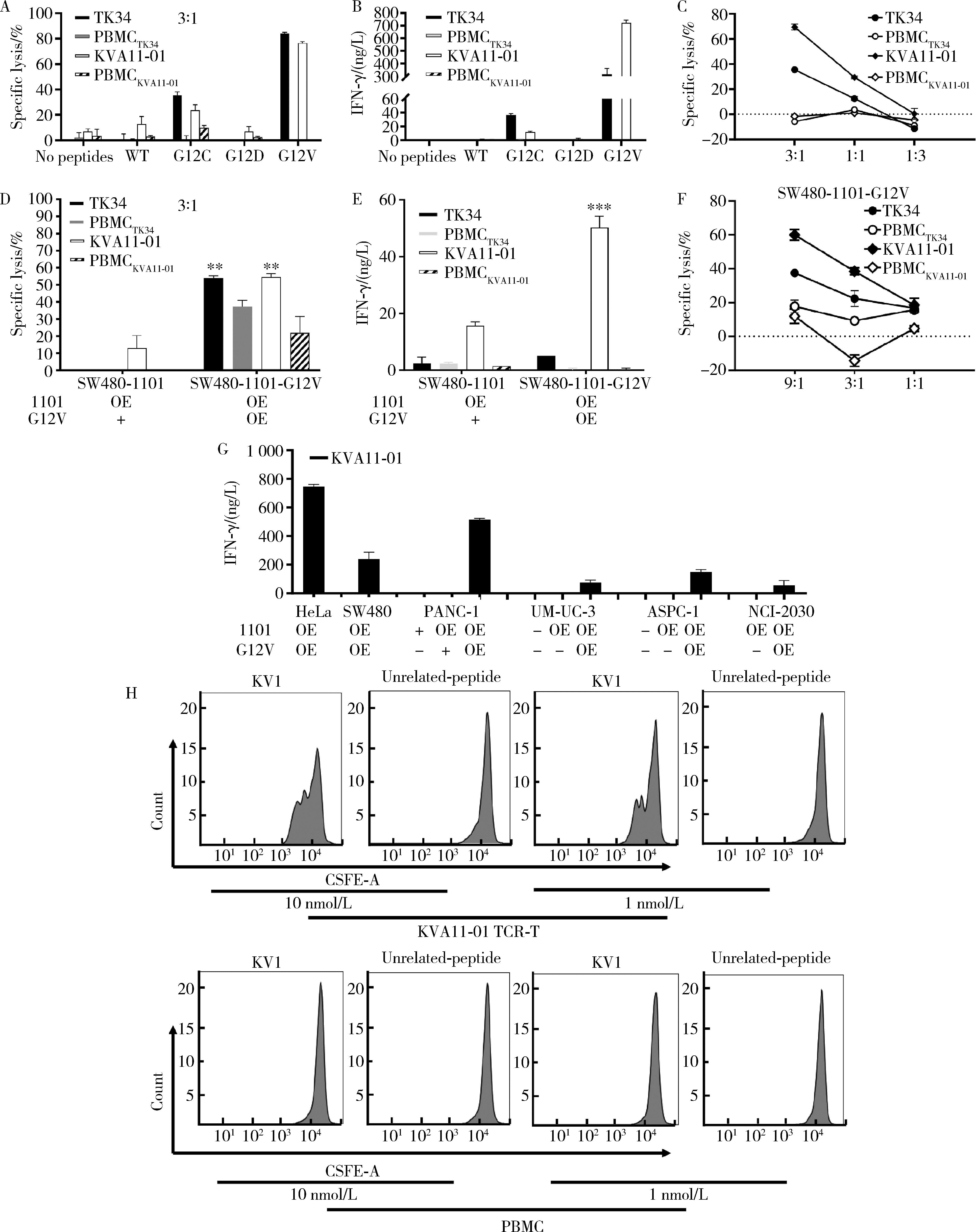
Journal of Peking University (Health Sciences) ›› 2022, Vol. 54 ›› Issue (5): 884-895. doi: 10.19723/j.issn.1671-167X.2022.05.016
Previous Articles Next Articles
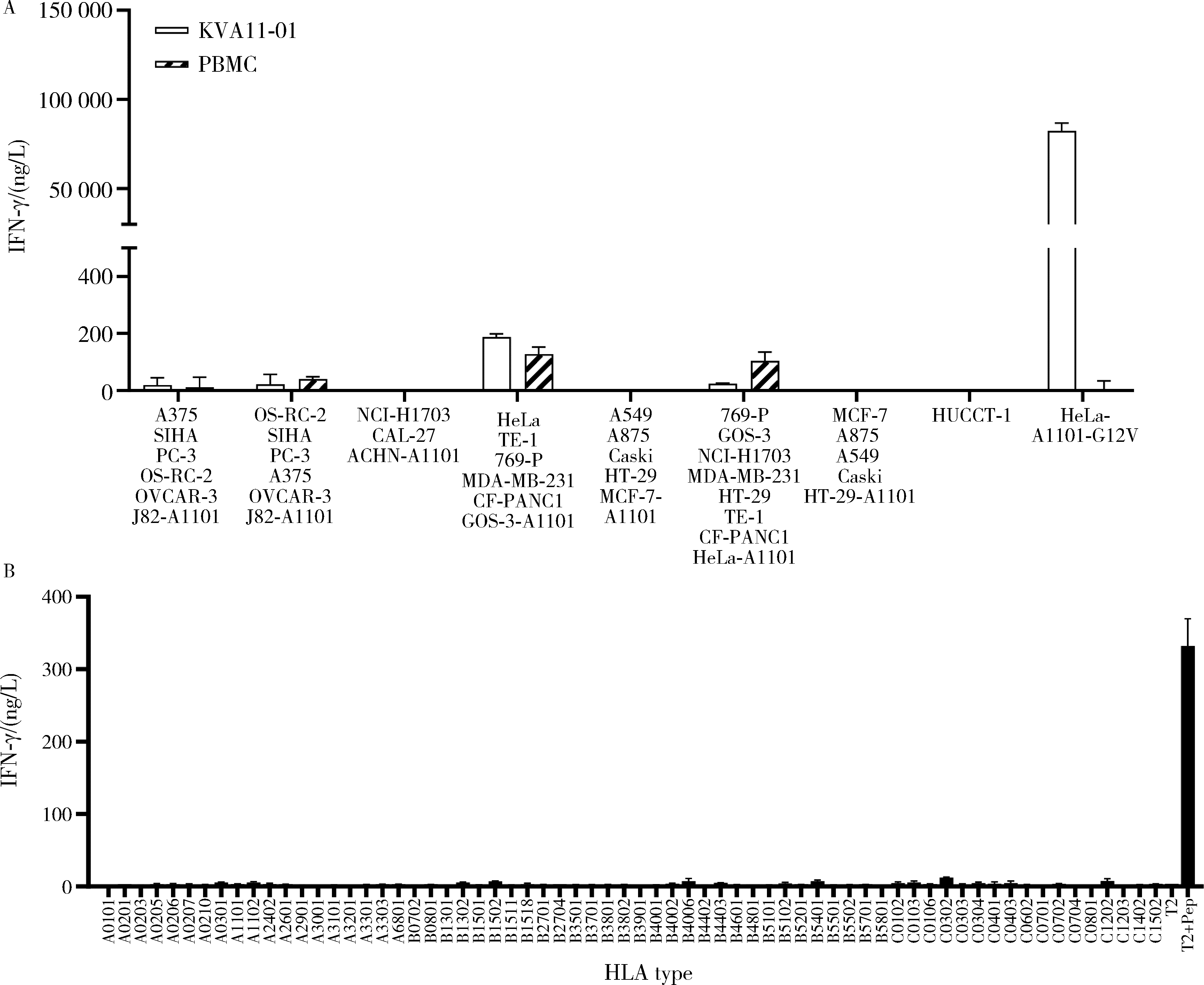
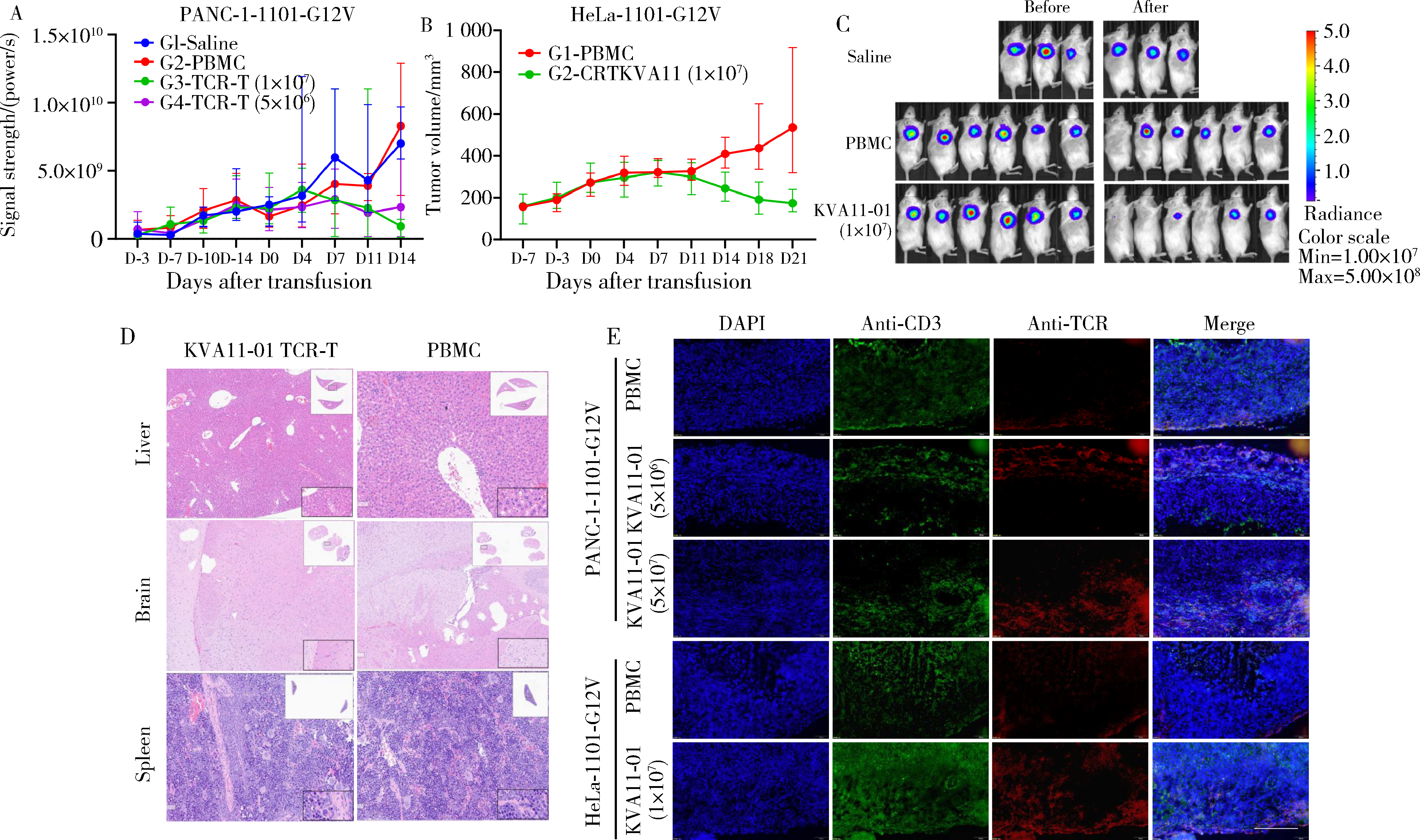
Preclinical study of T cell receptor specifically reactive with KRAS G12V mutation in the treatment of malignant tumors
Xiao-jing CHENG1,Dong JIANG2,Lian-hai ZHANG1,Jiang-hua WANG2,Ya-zhen LI2,Jia-hui ZHAI2,Bao-qi YAN2,Lu-lu ZHANG2,Xing-wang XIE2,*( ),Zi-yu LI1,*(
),Zi-yu LI1,*( ),Jia-fu JI1,*(
),Jia-fu JI1,*( )
)
- 1. Department of Gastrointestinal Cancer Center, Key Laboratory of Carcinogenesis and Translational Research, Ministry of Education; Laboratory of Genetics, Peking University Cancer Hospital & Institute, Beijing 100142, China
2. Beijing CorreGene Biotechnology Co., Ltd., Beijing 100094, China
CLC Number:
- R730.51
| 1 | Bos JL . Ras oncogenes in human cancer: A review[J]. Cancer Res, 1989, 49 (17): 4682- 4689. |
| 2 |
Singh H , Longo DL , Chabner BA . Improving prospects for targeting RAS[J]. J Clin Oncol, 2015, 33 (31): 3650- 3659.
doi: 10.1200/JCO.2015.62.1052 |
| 3 | Hobbs GA , Der CJ , Rossman KL . RAS isoforms and mutations in cancer at a glance[J]. J Cell Sci, 2016, 129 (7): 1287- 1292. |
| 4 |
Windon AL , Loaiza-Bonilla A , Jensen CE , et al. A KRAS wild type mutational status confers a survival advantage in pancreatic ductal adenocarcinoma[J]. J Gastrointest Oncol, 2018, 9 (1): 1- 10.
doi: 10.21037/jgo.2017.10.14 |
| 5 |
Shin SH , Kim SC , Hong SM , et al. Genetic alterations of K-ras, p53, c-erbB-2, and DPC4 in pancreatic ductal adenocarcinoma and their correlation with patient survival[J]. Pancreas, 2013, 42 (2): 216- 222.
doi: 10.1097/MPA.0b013e31825b6ab0 |
| 6 |
Bournet B , Muscari F , Buscail C , et al. KRAS G12D mutation subtype is a prognostic factor for advanced pancreatic adenocarcinoma[J]. Clin Transl Gastroenterol, 2016, 7 (3): e157.
doi: 10.1038/ctg.2016.18 |
| 7 |
Haas M , Ormanns S , Baechmann S , et al. Extended RAS analysis and correlation with overall survival in advanced pancreatic cancer[J]. Br J Cancer, 2017, 116 (11): 1462- 1469.
doi: 10.1038/bjc.2017.115 |
| 8 |
Marabese M , Ganzinelli M , Garassino MC , et al. KRAS mutations affect prognosis of non-small-cell lung cancer patients treated with first-line platinum containing chemotherapy[J]. Oncotarget, 2015, 6 (32): 34014- 34022.
doi: 10.18632/oncotarget.5607 |
| 9 |
Laghi L , Orbetegli O , Bianchi P , et al. Common occurrence of multiple KRAS mutations in pancreatic cancers with associated precursor lesions and in biliary cancers[J]. Oncogene, 2002, 21 (27): 4301- 4306.
doi: 10.1038/sj.onc.1205533 |
| 10 | Russo AL , Borger DR , Szymonifka J , et al. Mutational analysis and clinical correlation of metastatic colorectal cancer[J]. Can-cer, 2014, 120 (10): 1482- 1490. |
| 11 |
Tran E , Ahmadzadeh M , Lu YC , et al. Immunogenicity of soma-tic mutations in human gastrointestinal cancers[J]. Science, 2015, 350 (6266): 1387- 1390.
doi: 10.1126/science.aad1253 |
| 12 |
Chatani PD , Yang JC . Mutated RAS. Targeting the "untargetable" with T cells[J]. Clin Cancer Res, 2020, 26 (3): 537- 544.
doi: 10.1158/1078-0432.CCR-19-2138 |
| 13 |
Tran E , Robbins PF , Lu YC , et al. T-cell transfer therapy targeting mutant KRAS in cancer[J]. N Engl J Med, 2016, 375 (23): 2255- 2262.
doi: 10.1056/NEJMoa1609279 |
| 14 |
Leidner R , Sanjuan Silva N , Huang H , et al. Neoantigen T-cell receptor gene therapy in pancreatic cancer[J]. N Engl J Med, 2022, 386 (22): 2112- 2119.
doi: 10.1056/NEJMoa2119662 |
| 15 | Wang QJ , Yu Z , Griffith K , et al. Identification of T-cell receptors targeting KRAS-mutated human tumors[J]. Cancer Immunol Res, 2015, 4 (3): 204- 214. |
| 16 |
Sim MJW , Lu J , Spencer M , et al. High-affinity oligoclonal TCRs define effective adoptive T cell therapy targeting mutant KRAS-G12D[J]. Proc Natl Acad Sci USA, 2020, 117 (23): 12826- 12835.
doi: 10.1073/pnas.1921964117 |
| 17 |
Cohen CJ , Zhao Y , Zheng Z , et al. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability[J]. Cancer Res, 2006, 66 (17): 8878- 8886.
doi: 10.1158/0008-5472.CAN-06-1450 |
| 18 |
Morgan RA , Chinnasamy N , Abate-Daga D , et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy[J]. J Immunother, 2013, 36 (2): 133- 151.
doi: 10.1097/CJI.0b013e3182829903 |
| 19 |
Linette GP , Stadtmauer EA , Maus MV , et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma[J]. Blood, 2013, 122 (6): 863- 871.
doi: 10.1182/blood-2013-03-490565 |
| 20 | Cameron BJ , Gerry AB , Dukes J , et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells[J]. Sci Transl Med, 2013, 5 (197): 103- 126. |
| 21 |
Suchin EJ , Langmuir PB , Palmer E , et al. Quantifying the frequency of alloreactive T cells in vivo: New answers to an old question[J]. J Immunol, 2001, 166 (2): 973- 981.
doi: 10.4049/jimmunol.166.2.973 |
| 22 |
Li J , Li W , Huang K , et al. Chimeric antigen receptor T cell (CAR-T) immunotherapy for solid tumors: lessons learned and strategies for moving forward[J]. J Hematol Oncol, 2018, 11 (1): 22.
doi: 10.1186/s13045-018-0568-6 |
| 23 | Idorn M , Skadborg SK , Kellermann L , et al. Chemokine receptor engineering of T cells with CXCR2 improves homing towards subcutaneous human melanomas in xenograft mouse model[J]. Oncoimmunology, 2018, 7 (8): e1450715. |
| 24 |
Draper LM , Kwong ML , Gros A , et al. Targeting of HPV-16+ epithelial cancer cells by TCR gene engineered T cells directed against E6[J]. Clin Cancer Res, 2015, 21 (19): 4431- 4439.
doi: 10.1158/1078-0432.CCR-14-3341 |
| 25 |
Karapetis CS , Khambata-Ford S , Jonker DJ , et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer[J]. N Engl J Med, 2008, 359 (17): 1757- 1765.
doi: 10.1056/NEJMoa0804385 |
| 26 |
Moore AR , Rosenberg SC , McCormick F , et al. RAS-targeted therapies: Is the undruggable drugged?[J]. Nat Rev Drug Discov, 2020, 19 (8): 533- 552.
doi: 10.1038/s41573-020-0068-6 |
| [1] | Zhicun LI, Tianyu WU, Lei LIANG, Yu FAN, Yisen MENG, Qian ZHANG. Risk factors analysis and nomogram model construction of postoperative pathological upgrade of prostate cancer patients with single core positive biopsy [J]. Journal of Peking University (Health Sciences), 2024, 56(5): 896-901. |
| [2] | Yuxuan TIAN,Mingjian RUAN,Yi LIU,Derun LI,Jingyun WU,Qi SHEN,Yu FAN,Jie JIN. Predictive effect of the dual-parametric MRI modified maximum diameter of the lesions with PI-RADS 4 and 5 on the clinically significant prostate cancer [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 567-574. |
| [3] | Kaifeng YAO,Mingjian RUAN,Derun LI,Yuxuan TIAN,Yuke CHEN,Yu FAN,Yi LIU. Diagnostic efficacy of targeted biopsy combined with regional systematic biopsy in prostate cancer in patients with PI-RADS 4-5 [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 575-581. |
| [4] | Junyong OU,Kunming NI,Lulin MA,Guoliang WANG,Ye YAN,Bin YANG,Gengwu LI,Haodong SONG,Min LU,Jianfei YE,Shudong ZHANG. Prognostic factors of patients with muscle invasive bladder cancer with intermediate-to-high risk prostate cancer [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 582-588. |
| [5] | Zi-xuan XUE,Shi-ying TANG,Min QIU,Cheng LIU,Xiao-jun TIAN,Min LU,Jing-han DONG,Lu-lin MA,Shu-dong ZHANG. Clinicopathologic features and prognosis of young renal tumors with tumor thrombus [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 802-811. |
| [6] | Yi LIU,Chang-wei YUAN,Jing-yun WU,Qi SHEN,Jiang-xi XIAO,Zheng ZHAO,Xiao-ying WANG,Xue-song LI,Zhi-song HE,Li-qun ZHOU. Diagnostic efficacy of prostate cancer using targeted biopsy with 6-core systematic biopsy for patients with PI-RADS 5 [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 812-817. |
| [7] | Min QIU,You-long ZONG,Bin-shuai WANG,Bin YANG,Chu-xiao XU,Zheng-hui SUN,Min LU,Lei ZHAO,Jian LU,Cheng LIU,Xiao-jun TIAN,Lu-lin MA. Treatment outcome of laparoscopic partial nephrectomy in patients with renal tumors of moderate to high complexity [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 833-837. |
| [8] | Chang-wei YUAN,De-run LI,Zhi-hua LI,Yi LIU,Gang-zhi SHAN,Xue-song LI,Li-qun ZHOU. Application of dynamic contrast enhanced status in multiparametric magnetic resonance imaging for prostatic cancer with PI-RADS 4 lesion [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 838-842. |
| [9] | Hui-li LIU,Yan-han LV,Xiao-xiao WANG,Min LI. Factors influencing the chronic post-surgical pain after laparoscopic surgery for elderly patients with urinary tract tumors [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 851-856. |
| [10] | Zhong CAO,Hong-bing CEN,Jian-hong ZHAO,Jun MEI,Ling-zhi QIN,Wei LIAO,Qi-lin AO. Expression and significance of INSM1 and SOX11 in pancreatic neuroendocrine tumor and solid pseudopapillary neoplasm [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 575-581. |
| [11] | Jian-xun MA,You-chen XIA,Bi LI,Hong-mei ZHAO,Yu-tao LEI,Xi BU. Choice of immediate breast reconstructive methods after modified radical mastectomy [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 612-618. |
| [12] | Yu-xiao WU,Yi-fan KANG,Qian-ying MAO,Zi-meng LI,Xiao-feng SHAN,Zhi-gang CAI. Application of methylene blue near-infrared fluorescence imaging for oral sentinel lymph node mapping in rats [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 684-688. |
| [13] | Shang XIE,Zhi-gang CAI,Xiao-feng SHAN. Application value of whole exon sequencing and immune related indicators in the precision treatment of oral squamous cell carcinoma [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 697-701. |
| [14] | Han LU,Jian-yun ZHANG,Rong YANG,Le XU,Qing-xiang LI,Yu-xing GUO,Chuan-bin GUO. Clinical factors affecting the prognosis of lower gingival squamous cell carcinoma [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 702-707. |
| [15] | Yun-jing ZHANG,Li-ying QIAO,Meng QI,Ying YAN,Wei-wei KANG,Guo-zhen LIU,Ming-yuan WANG,Yun-feng XI,Sheng-feng WANG. Development and validation of risk prediction model for new-onset cardiovascular diseases among breast cancer patients: Based on regional medical data of Inner Mongolia [J]. Journal of Peking University (Health Sciences), 2023, 55(3): 471-479. |
|
||