Journal of Peking University (Health Sciences) ›› 2025, Vol. 57 ›› Issue (6): 1032-1041. doi: 10.19723/j.issn.1671-167X.2025.06.004
Previous Articles Next Articles
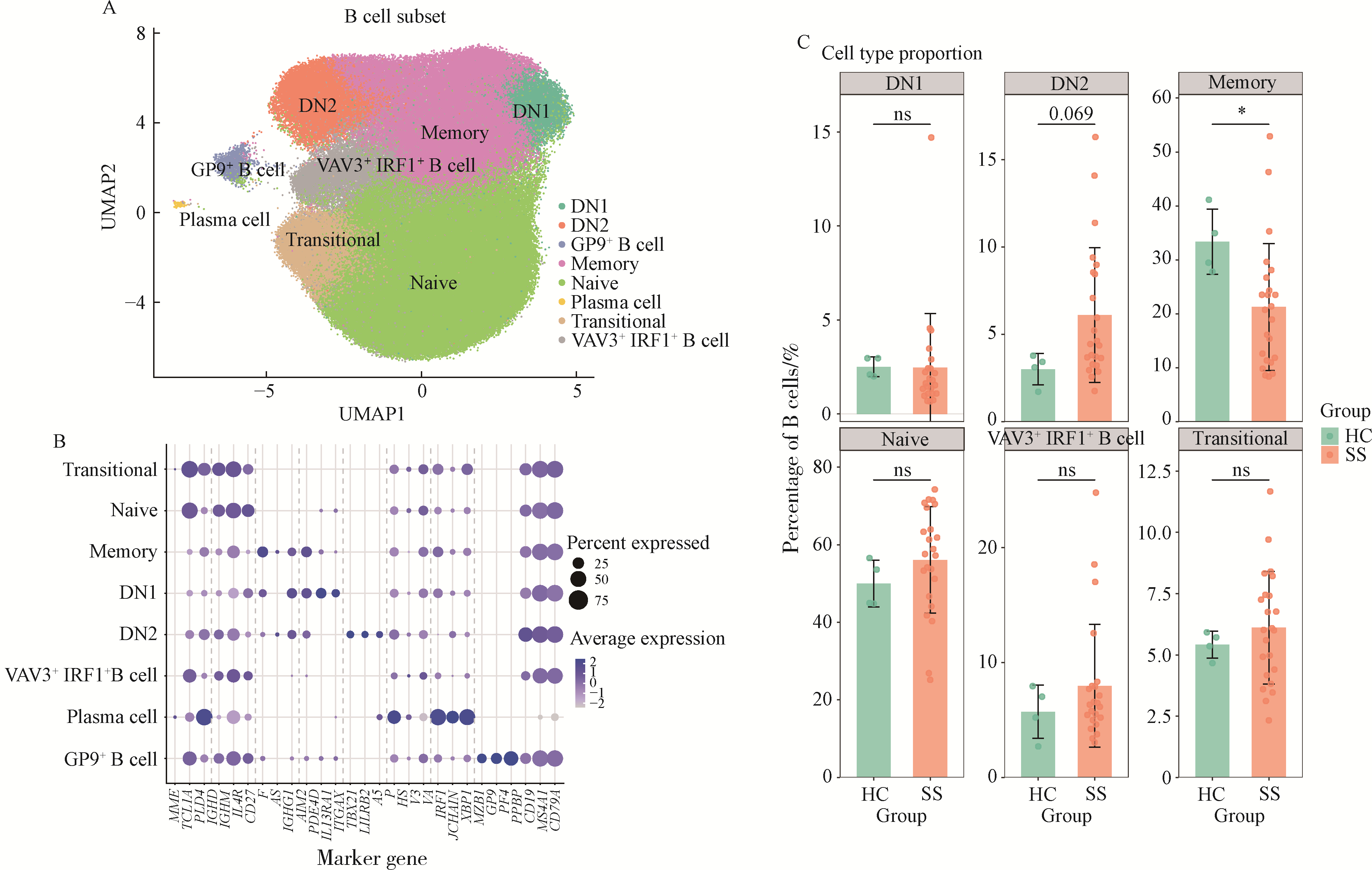
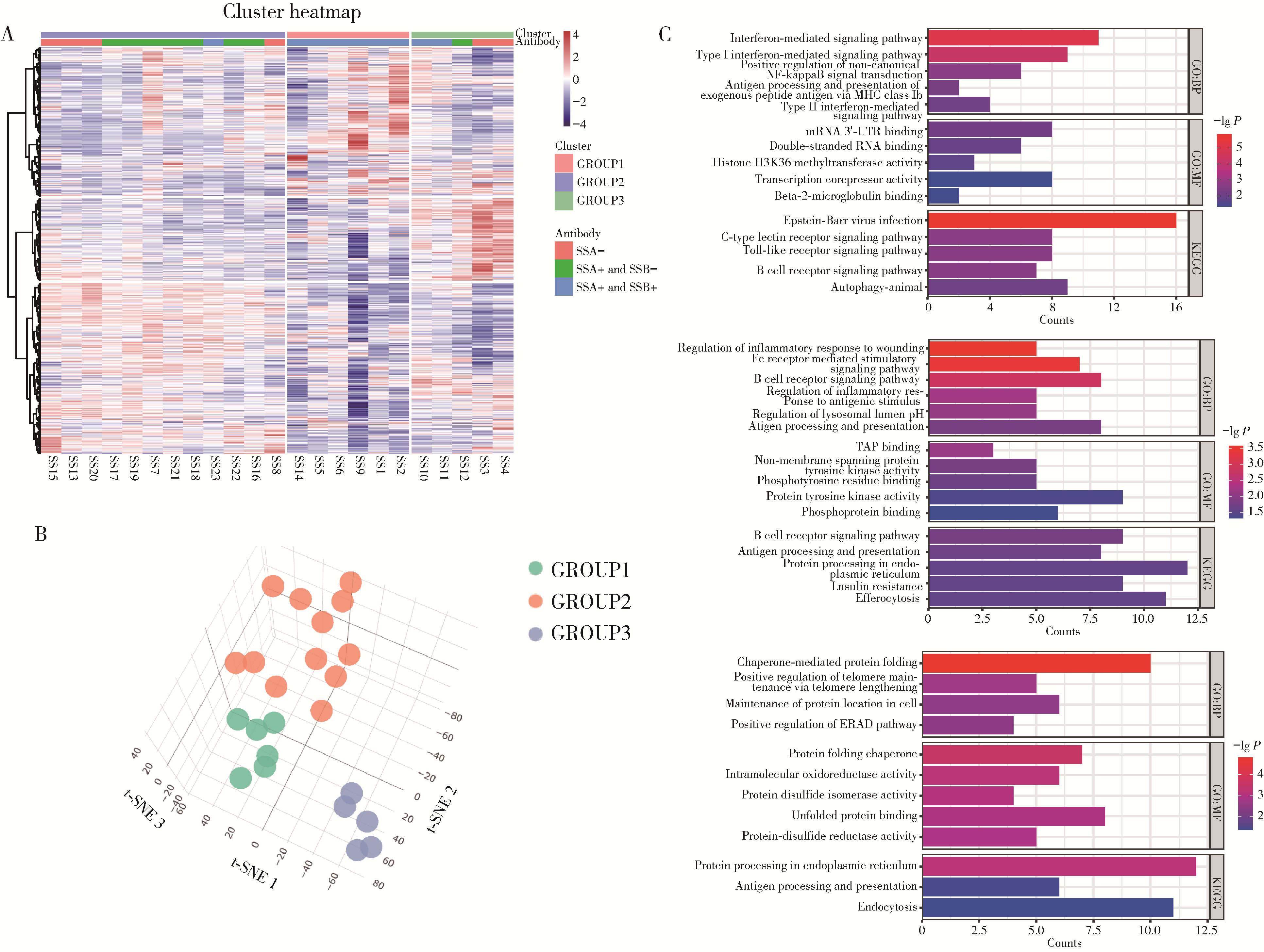
Single-cell RNA sequencing of B cells reveals molecular typing in Sjögren syndrome
Wenhao LIN, Yang XIE, Fangqing WANG, Shuying WANG, Xiangjun LIU, Fanlei HU*( ), Yuan JIA*(
), Yuan JIA*( )
)
- Department of Rheumatology and Immunology, Peking University People ' s Hospital, Beijing 100044, China
CLC Number:
- R593.2
| 1 |
中华医学会风湿病学分会. 干燥综合征诊断及治疗指南[J]. 中华风湿病学杂志, 2010, 14(11): 766- 768.
|
| 2 |
doi: 10.1038/nrrheum.2018.1 |
| 3 |
doi: 10.1016/j.immuni.2009.11.009 |
| 4 |
doi: 10.1002/art.42683 |
| 5 |
doi: 10.1186/s13073-023-01237-9 |
| 6 |
doi: 10.3389/fmolb.2023.1202371 |
| 7 |
doi: 10.1002/advs.202417593 |
| 8 |
doi: 10.1038/s41592-019-0619-0 |
| 9 |
|
| 10 |
doi: 10.1038/nbt.4096 |
| 11 |
doi: 10.1093/nar/gkac1000 |
| 12 |
doi: 10.1101/gr.1239303 |
| 13 |
doi: 10.1186/1471-2105-4-2 |
| 14 |
doi: 10.1016/S2665-9913(19)30042-6 |
| 15 |
doi: 10.1038/s41467-021-23472-7 |
| 16 |
doi: 10.1136/annrheumdis-2021-221604 |
| 17 |
doi: 10.1007/s10067-025-07409-9 |
| 18 |
doi: 10.3390/ijms24098385 |
| 19 |
doi: 10.1038/nri856 |
| 20 |
doi: 10.1186/s42358-024-00430-7 |
| 21 |
doi: 10.1016/j.canlet.2024.217045 |
| 22 |
doi: 10.2147/JIR.S413581 |
| 23 |
doi: 10.1038/374470a0 |
| 24 |
doi: 10.2147/JIR.S440263 |
| 25 |
doi: 10.1371/journal.pbio.3000252 |
| 26 |
杜晶晶, 董兴红, 高洁, 等. 原发性干燥综合征患者抗SSB与其他实验室参数相关性研究[J]. 检验医学与临床, 2023, 20(16): 2395- 2399.
|
| 27 |
doi: 10.1080/07853890.2022.2031272 |
| 28 |
doi: 10.1016/j.jtauto.2024.100252 |
| 29 |
doi: 10.1038/s41467-022-30651-7 |
| [1] | Yayun ZHAO, Mengfan NI, Xue LI, Bei WANG, Gong CHENG, Jing HE, Yuebo JIN. Clinical efficacy and safety of rituximab in treating renal injury in primary Sjögren syndrome [J]. Journal of Peking University (Health Sciences), 2025, 57(6): 1051-1060. |
| [2] | Zhao XIANG, Li YANG, Jing YANG. Untargeted metabolomics reveals differential serum metabolites and metabolic pathways in patients with primary Sjögren syndrome and thrombocytopenia [J]. Journal of Peking University (Health Sciences), 2025, 57(6): 1042-1050. |
| [3] | Yuan LIU, Guixiu SHI. Change from Sjögren syndrome to Sjögren disease [J]. Journal of Peking University (Health Sciences), 2025, 57(6): 1015-1017. |
| [4] | Lixiu ZHU, Renli CHEN, Sujuan ZHOU, Ye LIN, Yirong TANG, Zhen YE. Effect of aquaporin 5 on TLR4/MyD88/NF-κB signaling pathway in Sjögren syndrome rats [J]. Journal of Peking University (Health Sciences), 2025, 57(5): 875-883. |
| [5] | Xiang CAI,Rendong WANG,Shijia WANG,Ziqi REN,Qiuhong YU,Dongguo LI. Dynamic trajectory and cell communication of different cell clusters in malignant progression of glioblastoma [J]. Journal of Peking University (Health Sciences), 2024, 56(2): 199-206. |
|
||