Journal of Peking University (Health Sciences) ›› 2021, Vol. 53 ›› Issue (6): 1020-1025. doi: 10.19723/j.issn.1671-167X.2021.06.002
Previous Articles Next Articles
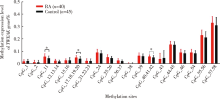
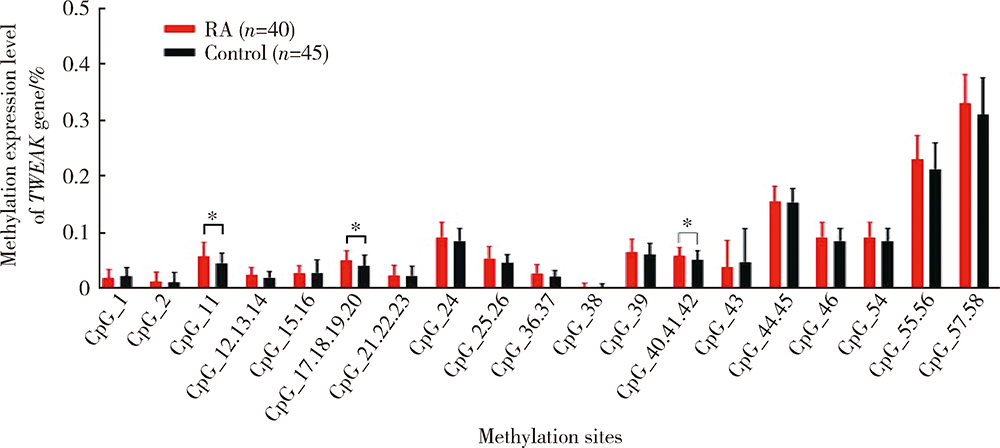
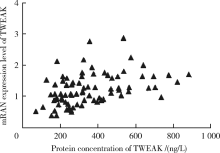
Methylation status and expression of TWEAK gene promoter region in peripheral blood of patients with rheumatoid arthritis
LOU Xue,LIAO Li,LI Xing-jun,WANG Nan,LIU Shuang,CUI Ruo-mei,XU Jian( )
)
- Department of Rheumatology and Immunology, First Affiliated Hospital of Kunming Medical University, Kunming 650032, China
CLC Number:
- R593.22
| [1] |
Firestein GS. Evolving concepts of rheumatoid arthritis[J]. Nature, 2003, 423(6937):356-361.
doi: 10.1038/nature01661 |
| [2] | Sánchez-Ramón S, López-Longo FJ, Carreño L. Interleukins network in rheumatoid arthritis pathophysiology: Beyond proinflammatory cytokines[J]. Reumatol Clin, 2011, 6(S3):S20-S24. |
| [3] |
Song X, Lin Q. Genomics, transcriptomics and proteomics to elucidate the pathogenesis of rheumatoid arthritis[J]. Rheumatol Int, 2017, 37(8):1257-1265.
doi: 10.1007/s00296-017-3732-3 |
| [4] |
Karami J, Aslani S, Jamshidi A, et al. Genetic implications in the pathogenesis of rheumatoid arthritis: An updated review[J]. Gene, 2019, 702:8-16.
doi: S0378-1119(19)30290-2 pmid: 30904715 |
| [5] |
Park J, Kwok S, Lim M, et al. TWEAK promotes osteoclastogenesis in rheumatoid arthritis[J]. Am J Pathol, 2013, 183(3):857-867.
doi: 10.1016/j.ajpath.2013.05.027 |
| [6] |
Chicheportiche Y, Chicheportiche R, Sizing I, et al. Proinflammatory activity of TWEAK on human dermal fibroblasts and synoviocytes: Blocking and enhancing effects of anti-TWEAK monoclonal antibodies[J]. Arthritis Res, 2002, 4(2):126-133.
pmid: 11879548 |
| [7] |
Karami J, Aslani S, Tahmasebi MN, et al. Epigenetics in rheumatoid arthritis; fibroblast-like synoviocytes as an emerging paradigm in the pathogenesis of the disease[J]. Immunol Cell Biol, 2020, 98(3):171-186.
doi: 10.1111/imcb.12311 pmid: 31856314 |
| [8] |
Singer BD. A practical guide to the measurement and analysis of DNA methylation[J]. Am J Respir Cell Mol Biol, 2019, 61(4):417-428.
doi: 10.1165/rcmb.2019-0150TR |
| [9] |
Hua XM, Wang J, Qian DM, et al. DNA methylation level of promoter region of activating transcription factor 5 in glioma[J]. J Zhejiang Univ Sci B, 2015, 16(9):757-762.
doi: 10.1631/jzus.B1500067 |
| [10] |
Fuso A, Raia T, Orticello M, et al. The complex interplay between DNA methylation and miRNAs in gene expression regulation[J]. Biochimie, 2020, 173:12-16.
doi: 10.1016/j.biochi.2020.02.006 |
| [11] | 中华医学会风湿病学分会. 类风湿关节炎诊断及治疗指南[J]. 中华风湿病学杂志, 2010(4):265-270. |
| [12] |
Schwartz N, Su L, Burkly LC, et al. Urinary TWEAK as a biomarker of lupus nephritis: A multicenter cohort study[J]. Arthritis Res Ther, 2009, 11(5):R143.
doi: 10.1186/ar2816 |
| [13] |
Xu W, Zhao Y, Liu Y. Role of the TWEAK/Fn14 pathway in autoimmune diseases[J]. Immunol Res, 2016, 64(1):44-50.
doi: 10.1007/s12026-015-8761-y |
| [14] |
Kamijo S, Nakajima A, Kamata K, et al. Involvement of TWEAK/Fn14 interaction in the synovial inflammation of RA[J]. Rheumatology (Oxford), 2008, 47(4):442-450.
doi: 10.1093/rheumatology/ken006 pmid: 18310134 |
| [15] | Bertin D, Stephan D, Khrestchatisky M, et al. Is TWEAK a biomarker for autoimmune/chronic inflammatory diseases[J]. Front Immunol, 2013(4):489. |
| [16] |
van Kuijk AW, Wijbrandts CA, Vinkenoog M, et al. TWEAK and its receptor Fn14 in the synovium of patients with rheumatoid arthritis compared to psoriatic arthritis and its response to tumour necrosis factor blockade[J]. Ann Rheum Dis, 2010, 69(1):301-304.
doi: 10.1136/ard.2008.090548 pmid: 19147618 |
| [17] |
Park MC, Chung SJ, Jung SJ, et al. Relationship of serum TWEAK level to cytokine level, disease activity, and response to anti-TNF treatment in patients with rheumatoid arthritis[J]. Scand J Rheumatol, 2008, 37(3):173-178.
doi: 10.1080/03009740801898608 pmid: 18465450 |
| [18] |
Dharmapatni A, Smith MD, Crotti TN, et al. TWEAK and Fn14 expression in the pathogenesis of joint inflammation and bone erosion in rheumatoid arthritis[J]. Arthritis Res Ther, 2011, 13(2):R51.
doi: 10.1186/ar3294 |
| [19] |
Maecker H, Varfolomeev E, Kischkel F, et al. TWEAK attenuates the transition from innate to adaptive immunity[J]. Cell, 2005, 123(5):931-944.
doi: 10.1016/j.cell.2005.09.022 |
| [20] |
Guo SC, Zhu Q, Jiang T, et al. Genome-wide DNA methylation patterns in CD4+ T cells from Chinese Han patients with rheumatoid arthritis[J]. Mod Rheumatol, 2017, 27(3):441-447.
doi: 10.1080/14397595.2016.1218595 |
| [21] |
Zhu H, Wu LF, Mo XB, et al. Rheumatoid arthritis-associated DNA methylation sites in peripheral blood mononuclear cells[J]. Ann Rheum Dis, 2019, 78(1):36-42.
doi: 10.1136/annrheumdis-2018-213970 |
| [1] | Dongwu LIU, Jie CHEN, Mingli GAO, Jing YU. Rheumatoid arthritis with Castleman-like histopathology in lymph nodes: A case report [J]. Journal of Peking University (Health Sciences), 2024, 56(5): 928-931. |
| [2] | Huina HUANG,Jing ZHAO,Xiangge ZHAO,Ziran BAI,Xia LI,Guan WANG. Regulatory effect of lactate on peripheral blood CD4+ T cell subsets in patients with rheumatoid arthritis [J]. Journal of Peking University (Health Sciences), 2024, 56(3): 519-525. |
| [3] | Xiaofei TANG,Yonghong LI,Qiuling DING,Zhuo SUN,Yang ZHANG,Yumei WANG,Meiyi TIAN,Jian LIU. Incidence and risk factors of deep vein thrombosis in patients with rheumatoid arthritis [J]. Journal of Peking University (Health Sciences), 2024, 56(2): 279-283. |
| [4] | Xue ZOU,Xiao-juan BAI,Li-qing ZHANG. Effectiveness of tofacitinib combined with iguratimod in the treatment of difficult-to-treat moderate-to-severe rheumatoid arthritis [J]. Journal of Peking University (Health Sciences), 2023, 55(6): 1013-1021. |
| [5] | Qi WU,Yue-ming CAI,Juan HE,Wen-di HUANG,Qing-wen WANG. Correlation between dyslipidemia and rheumatoid arthritis associated interstitial lung disease [J]. Journal of Peking University (Health Sciences), 2023, 55(6): 982-992. |
| [6] | Jing-feng ZHANG,Yin-ji JIN,Hui WEI,Zhong-qiang YAO,Jin-xia ZHAO. Correlation analysis between body mass index and clinical characteristics of rheumatoid arthritis [J]. Journal of Peking University (Health Sciences), 2023, 55(6): 993-999. |
| [7] | Yin-ji JIN,Lin SUN,Jin-xia ZHAO,Xiang-yuan LIU. Significance of IgA isotype of anti-v-raf murine sarcoma viral oncogene homologue B1 antibody in rheumatoid arthritis [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 631-635. |
| [8] | Wen-xin CAI,Shi-cheng LI,Yi-ming LIU,Ru-yu LIANG,Jing LI,Jian-ping GUO,Fan-lei HU,Xiao-lin SUN,Chun LI,Xu LIU,Hua YE,Li-zong DENG,Ru LI,Zhan-guo LI. A cross-sectional study on the clinical phenotypes of rheumatoid arthritis [J]. Journal of Peking University (Health Sciences), 2022, 54(6): 1068-1073. |
| [9] | Fang CHENG,Shao-ying YANG,Xing-xing FANG,Xuan WANG,Fu-tao ZHAO. Role of the CCL28-CCR10 pathway in monocyte migration in rheumatoid arthritis [J]. Journal of Peking University (Health Sciences), 2022, 54(6): 1074-1078. |
| [10] | Rui LIU,Jin-xia ZHAO,Liang YAN. Clinical characteristics of patients with rheumatoid arthritis complicated with venous thrombosis of lower extremities [J]. Journal of Peking University (Health Sciences), 2022, 54(6): 1079-1085. |
| [11] | Jing-feng ZHANG,Yin-ji JIN,Hui WEI,Zhong-qiang YAO,Jin-xia ZHAO. Cross-sectional study on quality of life and disease activity of rheumatoid arthritis patients [J]. Journal of Peking University (Health Sciences), 2022, 54(6): 1086-1093. |
| [12] | GAO Chao,CHEN Li-hong,WANG Li,YAO Hong,HUANG Xiao-wei,JIA Yu-bo,LIU Tian. Validation of the Pollard’s classification criteria (2010) for rheumatoid arthritis patients with fibromyalgia [J]. Journal of Peking University (Health Sciences), 2022, 54(2): 278-282. |
| [13] | ZHONG Hua,XU Li-ling,BAI Ming-xin,SU Yin. Effect of chemokines CXCL9 and CXCL10 on bone erosion in patients with rheumatoid arthritis [J]. Journal of Peking University (Health Sciences), 2021, 53(6): 1026-1031. |
| [14] | LUO Liang,HUO Wen-gang,ZHANG Qin,LI Chun. Clinical characteristics and risk factors of rheumatoid arthritis with ulcerative keratitis [J]. Journal of Peking University (Health Sciences), 2021, 53(6): 1032-1036. |
| [15] | ZHANG Lu,HU Xiao-hong,CHEN Cheng,CAI Yue-ming,WANG Qing-wen,ZHAO Jin-xia. Analysis of cervical instability and clinical characteristics in treatment-naive rheumatoid arthritis patients [J]. Journal of Peking University (Health Sciences), 2021, 53(6): 1049-1054. |
|
||