Journal of Peking University (Health Sciences) ›› 2024, Vol. 56 ›› Issue (5): 763-774. doi: 10.19723/j.issn.1671-167X.2024.05.003
Previous Articles Next Articles
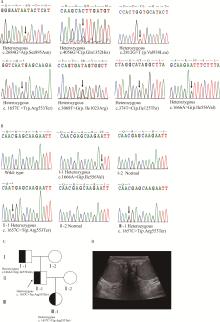
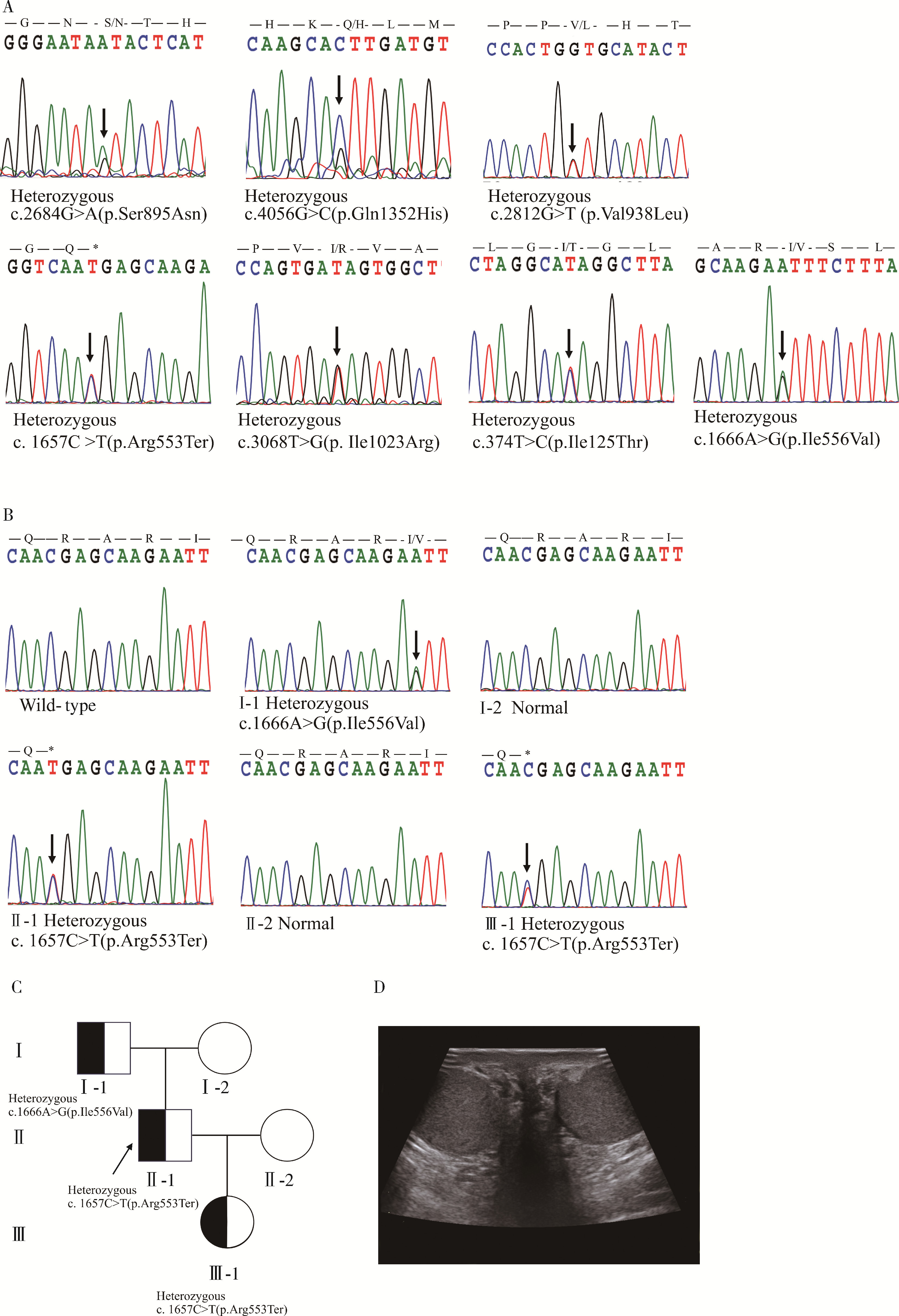
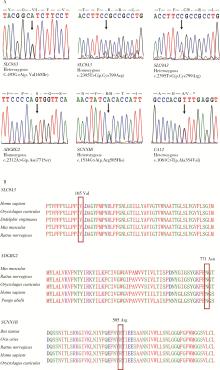
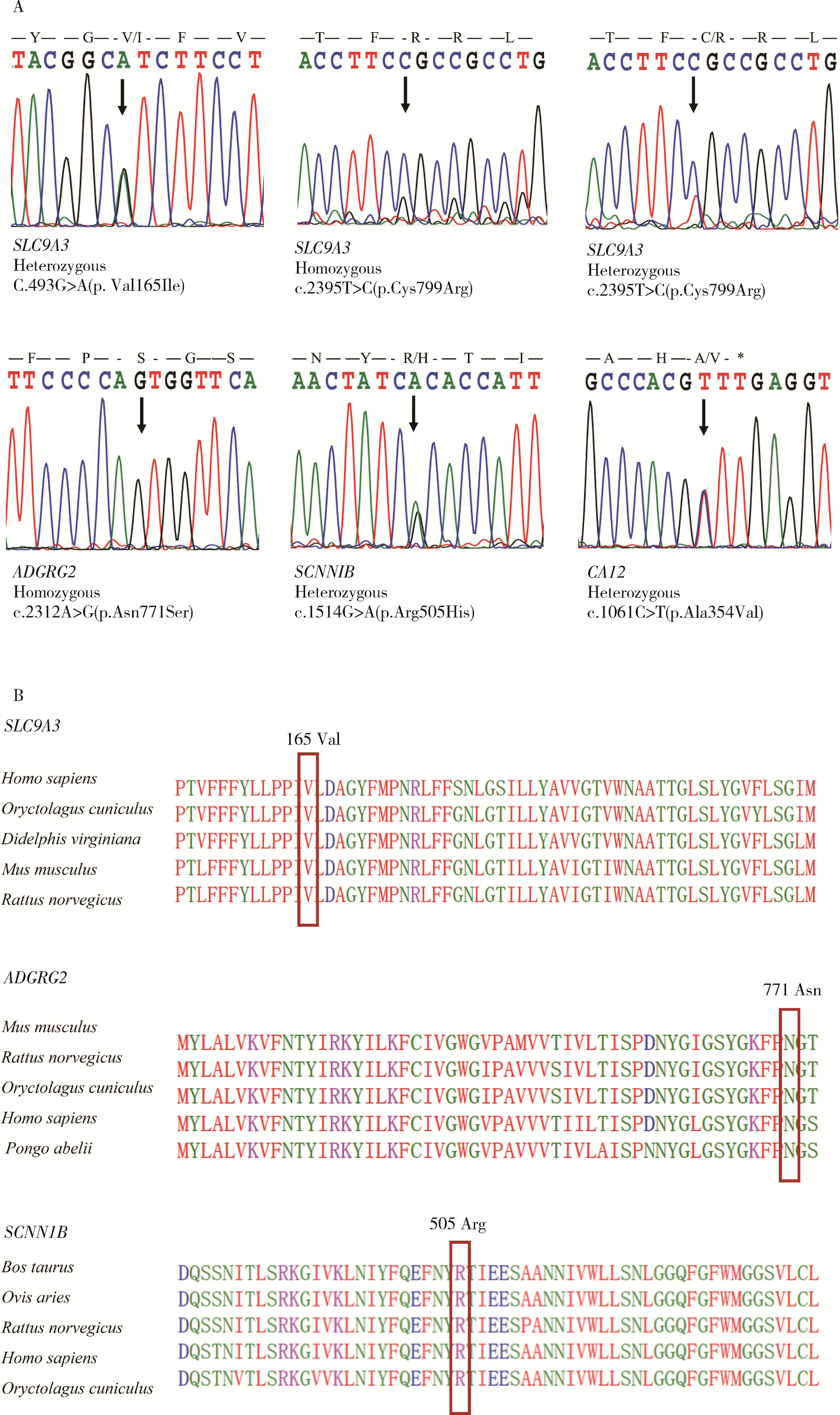
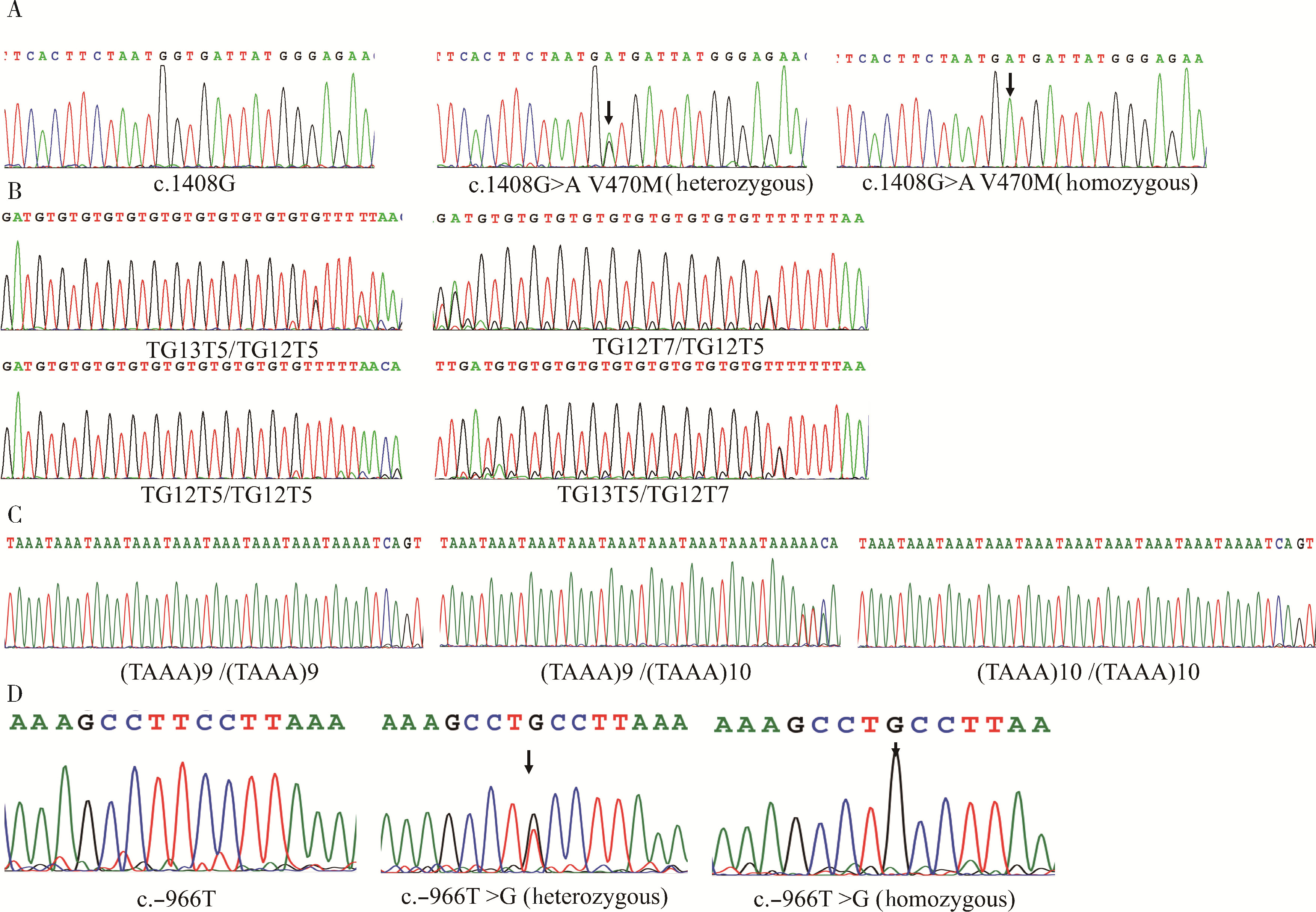
Detection of pathogenic gene mutations in thirteen cases of congenital bilateral absence of vas deferens infertility patients
Ying TANG, Yongbo ZHANG, Danhong WU, Yanhong LIN, Fenghua LAN*( )
)
- Fujian Provincial Key Laboratory of Transplant Biology, Fuzong Clinical Medical College of Fujian Medical University (The 900th Hospital of Joint Logistic Support Force, PLA), Fuzhou 350025, China
CLC Number:
- R394.1
| 1 |
Yang B , Lei C , Yang D , et al. Whole-exome sequencing identified CFTR variants in two consanguineous families in China[J]. Front Genet, 2021, 12, 631221.
doi: 10.3389/fgene.2021.631221 |
| 2 |
王宇刚, 马淑霞, 姚晓飞, 等. 男性不育伴精液量少的临床特点分析[J]. 生殖医学杂志, 2023, 32 (7): 997- 1002.
doi: 10.3969/j.issn.1004-3845.2023.07.005 |
| 3 |
Chillón M , Casals T , Mercier B , et al. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens[J]. N Engl J Med, 1995, 332 (22): 1475- 1480.
doi: 10.1056/NEJM199506013322204 |
| 4 |
Qu X , Li L , Cui C , et al. Correlation between CFTR variants and outcomes of ART in patients with CAVD in Central China[J]. Sci Rep, 2023, 13 (1): 64.
doi: 10.1038/s41598-022-26384-8 |
| 5 |
Wang H , An M , Liu Y , et al. Genetic diagnosis and sperm retrieval outcomes for Chinese patients with congenital bilateral absence of vas deferens[J]. Andrology, 2020, 8 (5): 1064- 1069.
doi: 10.1111/andr.12769 |
| 6 |
Fang J , Wang X , Sun X , et al. Congenital absence of the vas deferens with hypospadias or without hypospadias: Phenotypic findings and genetic considerations[J]. Front Genet, 2022, 13, 1035468.
doi: 10.3389/fgene.2022.1035468 |
| 7 |
Bieth E , Hamdi SM , Mieusset R . Genetics of the congenital absence of the vas deferens[J]. Hum Genet, 2021, 140 (1): 59- 76.
doi: 10.1007/s00439-020-02122-w |
| 8 |
Lu Y , Xie Y , Li M , et al. A novel ADGRG2 truncating variant associated with X-linked obstructive azoospermia in a large Chinese pedigree[J]. J Assist Reprod Genet, 2023, 40 (7): 1747- 1754.
doi: 10.1007/s10815-023-02839-3 |
| 9 | Wu YN , Chen KC , Wu CC , et al. SLC9A3 affects vas deferens development and associates with Taiwanese congenital bilateral absence of the vas deferens[J]. Biomed Res Int, 2019, 10, 3562719. |
| 10 |
Shen Y , Yue HX , Li FP , et al. SCNN1B and CA12 play vital roles in occurrence of congenital bilateral absence of vas deferens (CBAVD)[J]. Asian J Androl, 2019, 21 (5): 525- 527.
doi: 10.4103/aja.aja_112_18 |
| 11 |
Guo X , Liu K , Liu Y , et al. Clinical and genetic characteristics of cystic fibrosis in Chinese patients: A systemic review of reported cases[J]. Orphanet J Rare Dis, 2018, 13 (1): 224.
doi: 10.1186/s13023-018-0968-2 |
| 12 |
Souza PFN , Amaral JL , Bezerra LP , et al. ACE2-derived peptides interact with the RBD domain of SARS-CoV-2 spike glycoprotein, disrupting the interaction with the human ACE2 receptor[J]. J Biomol Struct Dyn, 2022, 40 (12): 5493- 5506.
doi: 10.1080/07391102.2020.1871415 |
| 13 | 顾怡栋, 邓学东, 程洪波, 等. CBAVD患者超声分型在穿刺取精方式选择中的应用价值[J]. 医学影像学杂志, 2019, 29 (9): 1621- 1625. |
| 14 |
Prins S , Corradi V , Sheppard DN , et al. Can two wrongs make a right? F508del-CFTR ion channel rescue by second-site mutations in its transmembrane domains[J]. J Biol Chem, 2022, 298, 101615.
doi: 10.1016/j.jbc.2022.101615 |
| 15 |
Gaikwad A , Khan S , Kadam S , et al. Cystic fibrosis transmembrane conductance regulator-related male infertility: Relevance of genetic testing & counselling in Indian population[J]. Indian J Med Res, 2020, 152 (6): 575- 583.
doi: 10.4103/ijmr.IJMR_906_18 |
| 16 | Li H , Lin L , Hu X , et al. Liver failure in a Chinese cystic fibrosis child with homozygous R553X mutation[J]. Front Pediatr, 2019, 20 (7): 36. |
| 17 |
Leung GKC , Ying D , Mak CCY , et al. CFTR founder mutation causes protein trafficking defects in Chinese patients with cystic fibrosis[J]. Mol Genet Genomic Med, 2017, 5 (1): 40- 49.
doi: 10.1002/mgg3.258 |
| 18 |
Luo S , Feng J , Zhang Y , et al. Mutation analysis of the cystic fibrosis transmembrane conductance regulator gene in Chinese congenital absence of vas deferens patients[J]. Gene, 2021, 765, 145045.
doi: 10.1016/j.gene.2020.145045 |
| 19 |
Tadaka S , Hishinuma E , Komaki S , et al. jMorp updates in 2020:Large enhancement of multi-omics data resources on the general Japanese population[J]. Nucleic Acids Res, 2021, 49 (D1): D536- D544.
doi: 10.1093/nar/gkaa1034 |
| 20 |
Le VS , Tran KT , Bui H TP , et al. A Vietnamese human genetic variation database[J]. Hum Mutat, 2019, 40 (10): 1664- 1675.
doi: 10.1002/humu.23835 |
| 21 |
Gaikwad A , Khan S , Kadam S , et al. The CFTR gene mild variants poly-T, TG repeats and M470V detection in Indian men with congenital bilateral absence of vas deferens[J]. Andrologia, 2018, 50 (2): e12858.
doi: 10.1111/and.12858 |
| 22 |
Casals T , Bassas L , Egozcue S , et al. Heterogeneity for mutations in the CFTR gene and clinical correlations in patients with con-genital absence of the vas deferens[J]. Hum Reprod, 2000, 15 (7): 1476- 1483.
doi: 10.1093/humrep/15.7.1476 |
| 23 |
Grangeia A , Niel F , Carvalho F , et al. Characterization of cystic fibrosis conductance transmembrane regulator gene mutations and IVS8 poly(T) variants in Portuguese patients with congenital absence of the vas deferens[J]. Hum Reprod, 2004, 19 (11): 2502- 2508.
doi: 10.1093/humrep/deh462 |
| 24 |
Fathy M , Ramzy T , Elmonem MA , et al. Molecular screening of CFTR gene in Egyptian patients with congenital bilateral absence of the vas deferens: A preliminary study[J]. Andrologia, 2016, 48 (10): 1307- 1312.
doi: 10.1111/and.12563 |
| 25 |
Cheng H , Yang S , Meng Q , et al. Genetic analysis and intracytoplasmic sperm injection outcomes of Chinese patients with congenital bilateral absence of vas deferens[J]. J Assist Reprod Genet, 2022, 39 (3): 719- 728.
doi: 10.1007/s10815-022-02417-z |
| 26 |
Yuan P , Liang ZK , Liang H , et al. Expanding the phenotypic and genetic spectrum of Chinese patients with congenital absence of vas deferens bearing CFTR and ADGRG2 alleles[J]. Andrology, 2019, 7 (3): 329- 340.
doi: 10.1111/andr.12592 |
| 27 |
Ni WH , Jiang L , Fei QJ , et al. The CFTR polymorphisms poly-T, TG-repeats and M470V in Chinese males with congenital bila-teral absence of the vas deferens[J]. Asian J Androl, 2012, 14 (5): 687- 690.
doi: 10.1038/aja.2012.43 |
| 28 |
Tosco A , Castaldo A , Colombo C , et al. Clinical outcomes of a large cohort of individuals with the F508del/5T; TG12 CFTR ge-notype[J]. J Cyst Fibros, 2022, 21 (5): 850- 855.
doi: 10.1016/j.jcf.2022.04.020 |
| 29 |
Cuppens H , Lin W , Jaspers M , et al. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation[J]. J Clin Invest, 1998, 101 (2): 487- 496.
doi: 10.1172/JCI639 |
| 30 |
Nykamp K , Truty R , Riethmaier D , et al. Elucidating clinical phenotypic variability associated with the polyT tract and TG repeats in CFTR[J]. Hum Mutat, 2021, 42 (9): 1165- 1172.
doi: 10.1002/humu.24250 |
| 31 |
Kerschner JL , Meckler F , Coatti GC , et al. The impact of geno-mic distance on enhancer-promoter interactions at the CFTR locus[J]. J Cell Mol Med, 2024, 28 (4): e18142.
doi: 10.1111/jcmm.18142 |
| 32 |
Verlingue C , Vuillaumier S , Mercier B , et al. Absence of mutations in the interspecies conserved regions of the CFTR promoter region in cystic fibrosis (CF) and CF related patients[J]. J Med Genet, 1998, 35 (2): 137- 140.
doi: 10.1136/jmg.35.2.137 |
| 33 | Bai S , Du Q , Liu X , et al. The detection and significance of cys-tic fibrosis transmembrane conductance regulator gene promoter mutations in Chinese patients with congenital bilateral absence of the vas deferens[J]. Gene, 2018, 672 (25): 64- 71. |
| 34 |
Zhuo JL , Soleimani M , Li XC . New insights into the critical importance of intratubular Na(+)/H(+) exchanger 3 and its potential therapeutic implications in hypertension[J]. Curr Hypertens Rep, 2021, 23 (6): 34.
doi: 10.1007/s11906-021-01152-7 |
| 35 |
Wang YY , Lin YH , Wu YN , et al. Loss of SLC9A3 decreases CFTR protein and causes obstructed azoospermia in mice[J]. PLoS Genet, 2017, 13 (4): e1006715.
doi: 10.1371/journal.pgen.1006715 |
| 36 |
Karacan Küçükali G , çetinkaya S , Tunç G , et al. Clinical management in systemic type pseudohypoaldosteronism due to SCNN1B variant and literature review[J]. J Clin Res Pediatr Endocrinol, 2021, 13 (4): 446- 451.
doi: 10.4274/jcrpe.galenos.2020.2020.0107 |
| 37 |
Ideozu JE , Liu M , Riley-Gillis BM , et al. Diversity of CFTR variants across ancestries characterized using 454 727 UK biobank whole exome sequences[J]. Genome Med, 2024, 16 (1): 43.
doi: 10.1186/s13073-024-01316-5 |
| 38 |
Ni Q , Chen X , Zhang P , et al. Systematic estimation of cystic fibrosis prevalence in Chinese and genetic spectrum comparison to Caucasians[J]. Orphanet J Rare Dis, 2022, 17 (1): 129.
doi: 10.1186/s13023-022-02279-9 |
| [1] | Enci XUE, Xi CHEN, Xueheng WANG, Siyue WANG, Mengying WANG, Jin LI, Xueying QIN, Yiqun WU, Nan LI, Jing LI, Zhibo ZHOU, Hongping ZHU, Tao WU, Dafang CHEN, Yonghua HU. Single nucleotide polymorphism heritability of non-syndromic cleft lip with or without cleft palate in Chinese population [J]. Journal of Peking University (Health Sciences), 2024, 56(5): 775-780. |
| [2] | WU Jun-yi,YU Miao,SUN Shi-chen,FAN Zhuang-zhuang,ZHENG Jing-lei,ZHANG Liu-tao,FENG Hai-lan,LIU Yang,HAN Dong. Detection of EDA gene mutation and phenotypic analysis in patients with hypohidrotic ectodermal dysplasia [J]. Journal of Peking University (Health Sciences), 2021, 53(1): 24-33. |
|
||