Journal of Peking University (Health Sciences) ›› 2025, Vol. 57 ›› Issue (4): 684-691. doi: 10.19723/j.issn.1671-167X.2025.04.009
Previous Articles Next Articles
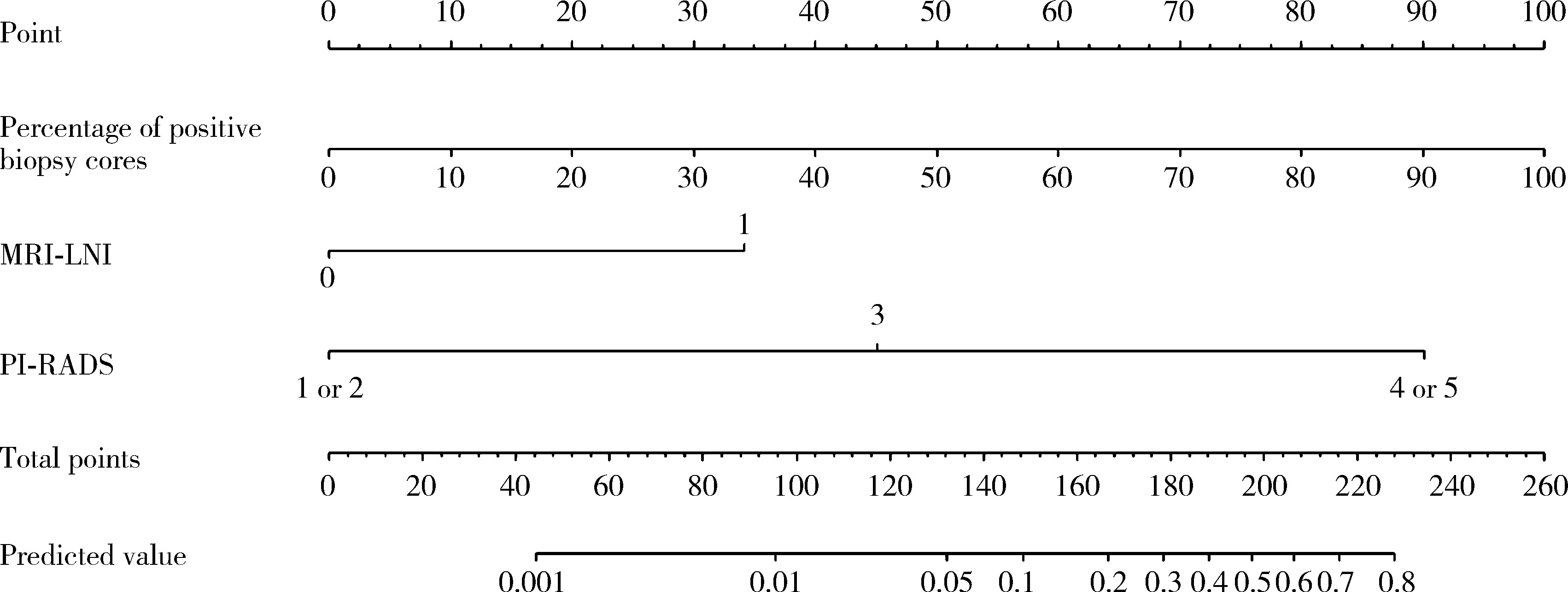
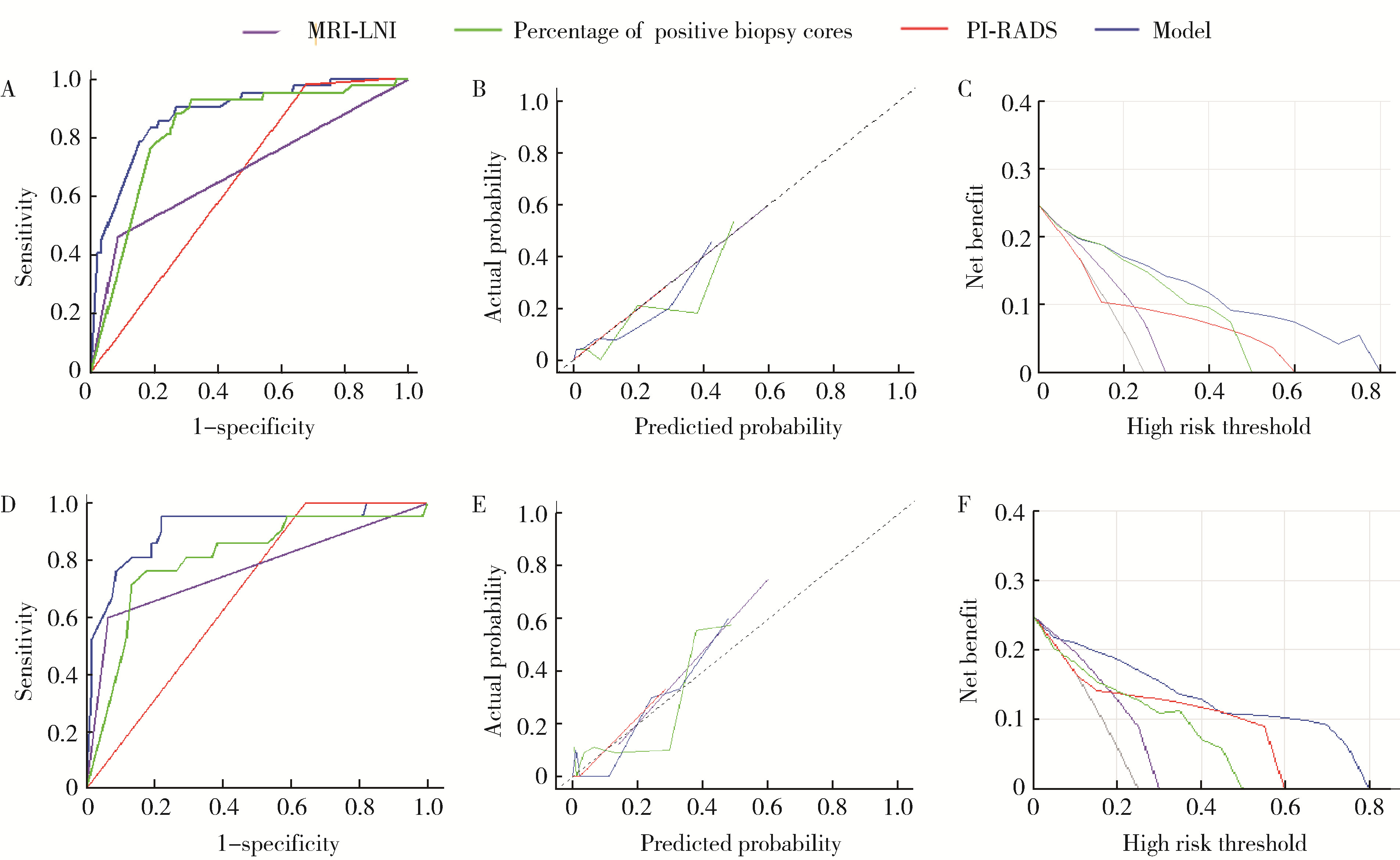
A preoperative prediction model for pelvic lymph node metastasis in prostate cancer: Integrating clinical characteristics and multiparametric MRI
Zeyuan WANG, Shuanbao YU, Haoke ZHENG, Jin TAO, Yafeng FAN, Xuepei ZHANG*( )
)
- Department of Urology, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450000, China
CLC Number:
- R737.25
| 1 |
|
| 2 |
|
| 3 |
|
| 4 |
|
| 5 |
|
| 6 |
|
| 7 |
|
| 8 |
|
| 9 |
|
| 10 |
|
| 11 |
|
| 12 |
|
| 13 |
|
| 14 |
|
| 15 |
|
| 16 |
|
| 17 |
|
| 18 |
|
| 19 |
|
| 20 |
|
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|
| 25 |
|
| 26 |
|
| 27 |
|
| 28 |
|
| [1] | Jiaxin NING, Haoran WANG, Shuhang LUO, Jibo JING, Jianye WANG, Huimin HOU, Ming LIU. Multi-omics analysis of the relationship between oxidative stress-related gene and prostate cancer [J]. Journal of Peking University (Health Sciences), 2025, 57(4): 633-643. |
| [2] | Yuan NING, Xiaoying ZHANG, Xue LI, Yuan LI, Jing HE, Yuebo JIN. Sjögren disease complicated by primary breast lymphoma: A case report [J]. Journal of Peking University (Health Sciences), 2025, 57(4): 808-811. |
| [3] | Zhicun LI, Tianyu WU, Lei LIANG, Yu FAN, Yisen MENG, Qian ZHANG. Risk factors analysis and nomogram model construction of postoperative pathological upgrade of prostate cancer patients with single core positive biopsy [J]. Journal of Peking University (Health Sciences), 2024, 56(5): 896-901. |
| [4] | Yuxuan TIAN,Mingjian RUAN,Yi LIU,Derun LI,Jingyun WU,Qi SHEN,Yu FAN,Jie JIN. Predictive effect of the dual-parametric MRI modified maximum diameter of the lesions with PI-RADS 4 and 5 on the clinically significant prostate cancer [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 567-574. |
| [5] | Kaifeng YAO,Mingjian RUAN,Derun LI,Yuxuan TIAN,Yuke CHEN,Yu FAN,Yi LIU. Diagnostic efficacy of targeted biopsy combined with regional systematic biopsy in prostate cancer in patients with PI-RADS 4-5 [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 575-581. |
| [6] | Junyong OU,Kunming NI,Lulin MA,Guoliang WANG,Ye YAN,Bin YANG,Gengwu LI,Haodong SONG,Min LU,Jianfei YE,Shudong ZHANG. Prognostic factors of patients with muscle invasive bladder cancer with intermediate-to-high risk prostate cancer [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 582-588. |
| [7] | Yi LIU,Chang-wei YUAN,Jing-yun WU,Qi SHEN,Jiang-xi XIAO,Zheng ZHAO,Xiao-ying WANG,Xue-song LI,Zhi-song HE,Li-qun ZHOU. Diagnostic efficacy of prostate cancer using targeted biopsy with 6-core systematic biopsy for patients with PI-RADS 5 [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 812-817. |
| [8] | Chang-wei YUAN,De-run LI,Zhi-hua LI,Yi LIU,Gang-zhi SHAN,Xue-song LI,Li-qun ZHOU. Application of dynamic contrast enhanced status in multiparametric magnetic resonance imaging for prostatic cancer with PI-RADS 4 lesion [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 838-842. |
| [9] | Yan XIONG,Xin LI,Li LIANG,Dong LI,Li-min YAN,Xue-ying LI,Ji-ting DI,Ting LI. Evaluation of accuracy of pathological diagnosis based on thyroid core needle biopsy [J]. Journal of Peking University (Health Sciences), 2023, 55(2): 234-242. |
| [10] | Dan-feng ZHENG,Jun-yu LI,Jia-xi LI,Ying-shuang ZHANG,Yan-feng ZHONG,Miao YU. Pathologic features of paraspinal muscle biopsies in patients with adolescent idiopathic scoliosis [J]. Journal of Peking University (Health Sciences), 2023, 55(2): 283-291. |
| [11] | ZHOU Guang-ping,ZHOU Qian-yun,ZHU Ji-hong. A case report of TAFRO syndrome [J]. Journal of Peking University (Health Sciences), 2021, 53(4): 814-817. |
| [12] | ZHANG Lei,LI Guo-liang,DANG Zong-hui, ,A yong,WU Ling-jie,LIU Li-jun. Analysis of bleeding risk in percutaneous renal biopsy in Tibet [J]. Journal of Peking University (Health Sciences), 2021, 53(2): 298-301. |
| [13] | WANG Ying-chun,HUANG Yong-hui,CHANG Hong,YAO Wei,YAN Xiu-e,LI Ke,ZHANG Yao-peng,ZHENG Wei. Characteristics of benign and malignant lesions of ampullary polyps and the accuracy of forceps biopsy [J]. Journal of Peking University (Health Sciences), 2021, 53(1): 204-209. |
| [14] | Ying-chao WU,Yun-long CAI,Long RONG,Ji-xin ZHANG,Jin LIU,Xin WANG. Characteristics of lymph node metastasis and evaluating the efficacy of endoscopic submucosal dissection in early gastric cancer [J]. Journal of Peking University (Health Sciences), 2020, 52(6): 1093-1097. |
| [15] | Yi LIU,Zhi-jian LIU,Qi SHEN,Jing-yun WU,Yu FAN,De-run LI,Wei YU,Zhi-song HE. A clinical analysis of 14 cases of prostatic stromal tumor of uncertain malignant potential [J]. Journal of Peking University (Health Sciences), 2020, 52(4): 621-624. |
|
||