北京大学学报(医学版) ›› 2024, Vol. 56 ›› Issue (2): 199-206. doi: 10.19723/j.issn.1671-167X.2024.02.001
• 论著 • 下一篇
胶质母细胞瘤恶性进展中不同细胞亚群的动态轨迹和细胞通讯网络
蔡祥1,王仁东1,王世佳1,任梓齐2,于秋红2,李冬果1,*( )
)
- 1. 首都医科大学生物医学工程学院智能医学工程学学系,北京 100069
2. 首都医科大学附属北京天坛医院高压氧科,北京 100070
Dynamic trajectory and cell communication of different cell clusters in malignant progression of glioblastoma
Xiang CAI1,Rendong WANG1,Shijia WANG1,Ziqi REN2,Qiuhong YU2,Dongguo LI1,*( )
)
- 1. Department of Intelligent Medical Engineering, School of Biomedical Engineering, Capital Medical University, Beijing 100069, China
2. Department of Hyperbaric Oxygen, Beijing Tiantan Hospital, Capital Medical University, Beijing 100070, China
摘要:
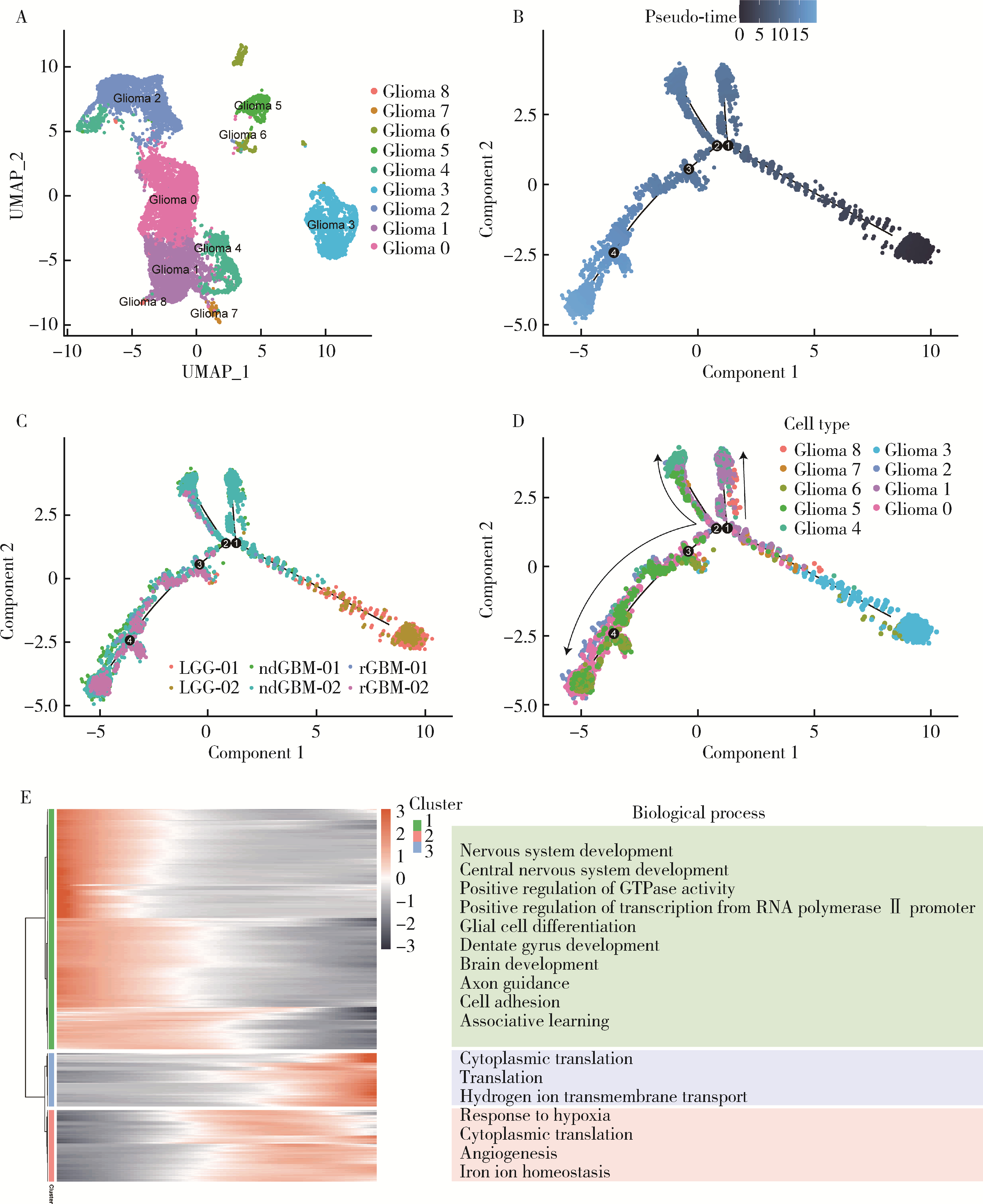
目的: 探索胶质母细胞瘤(glioblastoma, GBM)恶性进展过程中细胞亚群的动态轨迹以及免疫细胞亚群之间的通讯网络,结合GBM患者的转录组数据和临床信息,挖掘GBM恶性进展过程中的关键风险标志物,以期为该疾病的治疗和预后提供科学依据。方法: 基于单细胞测序数据分析方法,构建GBM恶性进展中的细胞亚群图谱,利用Monocle2技术构建GBM恶性进展中肿瘤细胞亚群的动态进展轨迹,基于基因富集分析,挖掘肿瘤细胞亚群随GBM恶性进展中显著变化的基因所富集的生物学过程,利用CellChat软件识别不同免疫细胞亚群间的复杂通讯网络,通过生存分析识别GBM恶性进展中影响患者预后的关键风险分子标记物。结果: 单细胞测序数据分析识别出6种不同的细胞类型,包括淋巴细胞、周细胞、少突神经胶质细胞、巨噬细胞、胶质瘤细胞、小胶质细胞,单细胞数据集中了27 151个细胞,其中包含3 881个来源于低级别胶质瘤患者的细胞,10 166个来源于新诊断GBM患者的细胞,13 104个来源于复发性胶质瘤患者的细胞。胶质瘤细胞亚群逆时序分析提示,胶质瘤细胞亚群在恶性进展中存在着明显的细胞异质性;免疫细胞亚群的细胞相互作用分析揭示,GBM恶性进展中不同免疫细胞亚群之间的通讯网络共识别出22条具有生物学意义的配体-受体对,涉及12条通路;生存分析识别出8个与GBM患者预后密切相关的基因,其中SERPINE1、COL6A1、SPP1、LTF、C1S、AEBP1、SAA1L是GBM患者的高风险基因,ABCC8是GBM患者的低风险基因。结论: 深入揭示了GBM恶性进展中胶质瘤细胞亚群的动态变化以及免疫细胞亚群之间的通讯模式,对于理解GBM的复杂生物学过程具有重要意义,为GBM的精准医疗和治疗决策提供了科学依据,也为GBM患者更准确的预后评估提供了新的线索。
中图分类号:
- R739.41
| 1 |
Lee E , Yong RL , Paddison P , et al.Comparison of glioblastoma (GBM) molecular classification methods[J].Semin Cancer Biol,2018,53,201-211.
doi: 10.1016/j.semcancer.2018.07.006 |
| 2 |
Hernández Martínez A , Madurga R , García-Romero N , et al.Unravelling glioblastoma heterogeneity by means of single-cell RNA sequencing[J].Cancer Lett,2022,527,66-79.
doi: 10.1016/j.canlet.2021.12.008 |
| 3 |
Suvà ML , Tirosh I .The glioma stem cell model in the era of single-cell genomics[J].Cancer Cell,2020,37(5):630-636.
doi: 10.1016/j.ccell.2020.04.001 |
| 4 |
Ochocka N , Segit P , Walentynowicz KA , et al.Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages[J].Nat Commun,2021,12(1):1151.
doi: 10.1038/s41467-021-21407-w |
| 5 |
Zhang H , Wang Y , Zhao Y , et al.PTX3 mediates the infiltration, migration, and inflammation-resolving-polarization of macrophages in glioblastoma[J].CNS Neurosci Ther,2022,28(11):1748-1766.
doi: 10.1111/cns.13913 |
| 6 |
Abdelfattah N , Kumar P , Wang C , et al.Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target[J].Nat Commun,2022,13(1):767.
doi: 10.1038/s41467-022-28372-y |
| 7 |
Hara T , Chanoch-Myers R , Mathewson ND , et al.Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma[J].Cancer Cell,2021,39(6):779-792. e11.
doi: 10.1016/j.ccell.2021.05.002 |
| 8 | Liu S , Wang Z , Zhu R , et al.Three differential expression analysis methods for RNA sequencing: Limma, EdgeR, DESeq2[J].J Vis Exp,2021,175,e62528. |
| 9 |
Korsunsky I , Millard N , Fan J , et al.Fast, sensitive and accurate integration of single-cell data with Harmony[J].Nat Methods,2019,16(12):1289-1296.
doi: 10.1038/s41592-019-0619-0 |
| 10 | Slovin S , Carissimo A , Panariello F , et al.Single-cell RNA sequencing analysis: A step-by-step overview[J].Methods Mol Biol,2021,2284,343-365. |
| 11 |
Butler A , Hoffman P , Smibert P , et al.Integrating single-cell transcriptomic data across different conditions, technologies, and species[J].Nat Biotechnol,2018,36(5):411-420.
doi: 10.1038/nbt.4096 |
| 12 |
Cao J , Spielmann M , Qiu X , et al.The single-cell transcriptional landscape of mammalian organogenesis[J].Nature,2019,566(7745):496-502.
doi: 10.1038/s41586-019-0969-x |
| 13 |
Dennis G , Jr Sherman BT , Hosack DA , et al.DAVID: Database for annotation, visualization, and integrated discovery[J].Genome Biol,2003,4(5):P3.
doi: 10.1186/gb-2003-4-5-p3 |
| 14 |
Jin S , Guerrero-Juarez CF , Zhang L , et al.Inference and analysis of cell-cell communication using CellChat[J].Nat Commun,2021,12(1):1088.
doi: 10.1038/s41467-021-21246-9 |
| 15 |
Lin J , Cai Y , Wang Z , et al.Novel biomarkers predict prognosis and drug-induced neuroendocrine differentiation in patients with prostate cancer[J].Front Endocrinol (Lausanne),2023,13,1005916.
doi: 10.3389/fendo.2022.1005916 |
| 16 |
Costa-Silva J , Domingues D , Lopes FM .RNA-Seq differential expression analysis: An extended review and a software tool[J].PLoS One,2017,12(12):e0190152.
doi: 10.1371/journal.pone.0190152 |
| 17 | D'Arrigo G , Leonardis D , Abd ElHafeez S , et al.Methods to analyse time-to-event data: The Kaplan-Meier survival curve[J].Oxid Med Cell Longev,2021,2021,2290120. |
| 18 |
DeCordova S , Shastri A , Tsolaki AG , et al.Molecular heterogeneity and immunosuppressive microenvironment in glioblastoma[J].Front Immunol,2020,11,1402.
doi: 10.3389/fimmu.2020.01402 |
| 19 |
Xiong Z , Yang Q , Li X .Effect of intra- and inter-tumoral heterogeneity on molecular characteristics of primary IDH-wild type glioblastoma revealed by single-cell analysis[J].CNS Neurosci Ther,2020,26(9):981-989.
doi: 10.1111/cns.13396 |
| 20 |
Põlajeva J , Sjösten AM , Lager N , et al.Mast cell accumulation in glioblastoma with a potential role for stem cell factor and chemokine CXCL12[J].PLoS One,2011,6(9):e25222.
doi: 10.1371/journal.pone.0025222 |
| 21 |
Xiong A , Zhang J , Chen Y , et al.Integrated single-cell transcriptomic analyses reveal that GPNMB-high macrophages promote PN-MES transition and impede T cell activation in GBM[J].EBioMedicine,2022,83,104239.
doi: 10.1016/j.ebiom.2022.104239 |
| 22 |
Chen P , Zhao D , Li J , et al.Symbiotic macrophage-glioma cell interactions reveal synthetic lethality in PTEN-null glioma[J].Cancer Cell,2019,35(6):868-884. e6.
doi: 10.1016/j.ccell.2019.05.003 |
| 23 |
Wu C , Qin C , Long W , et al.Tumor antigens and immune subtypes of glioblastoma: The fundamentals of mRNA vaccine and individualized immunotherapy development[J].J Big Data,2022,9(1):92.
doi: 10.1186/s40537-022-00643-x |
| [1] | 赵亮,孙丽芳,郑秀丽,刘静芳,郑蓉,张晗. 植入前囊胚滋养外胚层基因空间表达[J]. 北京大学学报(医学版), 2017, 49(6): 965-973. |
| [2] | 耿强, 陈红, 任景怡, 宋俊贤, 李素芳. 微颗粒携带microRNA-126在细胞间信号传递中的作用[J]. 北京大学学报(医学版), 2014, 46(6): 894-898. |
| [3] | 秦宇, 邓芙蓉, 魏红英, 韩丽敏, 许珺辉, 郭新彪. 纳米银材料中可溶性银离子对皮肤细胞间隙连接通讯的影响[J]. 北京大学学报(医学版), 2013, 45(03): 412-416. |
| [4] | 田新霞, 张云岗, 杜娟, 郑杰. 反义表皮生长因子受体cDNA转染对胶质母细胞瘤端粒酶活性的影响[J]. 北京大学学报(医学版), 2005, 37(3): 314-319. |
| [5] | 张志谦, 胡颖. 人肺上皮细胞、成纤维细胞、肺癌细胞间的异源间隙连接通讯[J]. 北京大学学报(医学版), 2005, 37(3): 306-309. |
|
||