北京大学学报(医学版) ›› 2019, Vol. 51 ›› Issue (4): 615-622. doi: 10.19723/j.issn.1671-167X.2019.04.003
基于长链非编码RNA的生物信息学分析构建膀胱癌预后模型并确定预后生物标志物
- 北京大学第三医院泌尿外科,北京 100191
Construction of prognostic model and identification of prognostic biomarkers based on the expression of long non-coding RNA in bladder cancer via bioinformatics
Fei-long YANG,Kai HONG,Guo-jiang ZHAO,Cheng LIU,Yi-meng SONG( ),Lu-lin MA(
),Lu-lin MA( )
)
- Department of Urology, Peking University Third Hospital, Beijing 100191, China
摘要:
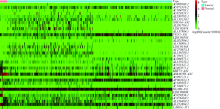
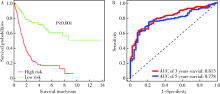
目的:构建基于长链非编码RNA(long non-coding RNA,lncRNA)的膀胱癌预后模型,并寻找预后生物标志物。方法:从癌症基因组图谱(The Cancer Genome Atlas,TCGA)数据库下载膀胱癌转录组及临床数据,Perl软件和R软件用于数据处理和分析。首先筛选差异表达lncRNA,继而对筛选结果进行单因素Cox回归分析以初步筛选与预后相关的lncRNA,再进一步用Lasso回归分析筛选影响预后的关键lncRNA,并运用多因素Cox回归分析构建预后模型。根据风险评分的中位数将患者分为高风险组和低风险组,运用Kaplan-Meier(K-M)生存分析、受试者接受特征(receiver operating characteristic,ROC)曲线和C指数对模型进行评价。此外,运用多因素Cox回归分析计算预后模型中各lncRNA的危险比和95%置信区间,并对差异有统计学意义的lncRNA进行K-M生存分析以确定预后生物标志物。结果:单因素Cox回归分析显示,在691个差异表达的lncRNA中, 35个可能与预后相关,其中23个经Lasso回归分析确认为影响预后的关键lncRNA。此外,K-M生存分析结果显示低风险组的总生存时间较高风险组长[(2.85±2.72)年vs. (1.58±1.51)年, P<0.001], ROC曲线显示3年生存率和5年生存率的曲线下面积分别为0.813和0.778,C指数为0.73。多因素Cox回归表明,23个关键lncRNA中有11个lncRNA差异有统计学意义,进一步的K-M生存分析表明,其中有3个lncRNA可能具有独立的预后价值,包括lncRNA AL589765.1(P = 0.004), AC023824.1(P = 0.022)和PKN2-AS1(P = 0.016)。结论:通过生物信息学分析,成功构建了基于23个lncRNA表达水平的膀胱癌预后模型,预测准确性中等,并确定了一个保护性预后生物标志物AL589765.1,以及两个不利的预后生物标志物AC023824.1和PKN2-AS1。
中图分类号:
- R737.11
| [1] | Bellmunt J, Orsola A, Leow JJ , et al. Bladder cancer: ESMO practice guidelines for diagnosis, treatment and follow-up[J]. Ann Oncol, 2014,25(Suppl 3):40-48. |
| [2] | Welty CJ, Sanford TH, Wright JL , et al. Thecancer of the bladder risk assessment (COBRA) score: estimating mortality after radical cystectomy[J]. Cancer, 2017,123(23):4574-4582. |
| [3] | 李吉, 刘裔道, 蚌凌青 . 肌层浸润性膀胱癌患者膀胱部分切除术结合放化疗的临床疗效及预后分析[J]. 临床泌尿外科杂志, 2017,32(10):767-770. |
| [4] | Esteller M . Non-coding RNAs in human disease[J]. Nat Rev Genet, 2011,12(12):861-874. |
| [5] | Martens-Uzunova ES, Bottcher R, Croce CM , et al. Long nonco-ding RNA in prostate, bladder, and kidney cancer[J]. Eur Urol, 2014,65(6):1140-1151. |
| [6] | Jin Y, Feng SJ, Qiu S , et al. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway[J]. Eur Rev Med Pharmacol Sci, 2017,21(14):3176-3184. |
| [7] | Gupta RA, Shah N, Wang KC , et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis[J]. Nature, 2010,464(7291):1071-1076. |
| [8] | Quinn JJ, Chang HY . Unique features of long non-coding RNA biogenesis and function[J]. Nat Rev Genet, 2016,17(1):47-62. |
| [9] | Djebali S, Davis CA, Merkel A , et al. Landscape of transcription in human cells[J]. Nature, 2012,489(7414):101-108. |
| [10] | Lee JT . Epigenetic regulation by long noncoding RNAs[J]. Science, 2012,338(6113):1435-1439. |
| [11] | Wang KC, Chang HY . Molecular mechanisms of long noncoding RNAs[J]. Mol Cell, 2011,43(6):904-914. |
| [12] | Liu D, Li Y, Luo G , et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2[J]. Cancer Lett, 2017,388:281-291. |
| [13] | Schmitt AM, Chang HY . Longnoncoding RNAs in cancer pathways[J]. Cancer Cell, 2016,29(4):452-463. |
| [14] | Yue B, Qiu S, Zhao S , et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis[J]. J Gastroenterol Hepatol, 2016,31(3):595-603. |
| [15] | Zhang S, Zhong G, He W , et al. lncRNA up-regulated in non-muscle invasive bladder cancer facilitates tumor growth and acts as a negative prognostic factor of recurrence[J]. J Urol, 2016,196(4):1270-1278. |
| [16] | Berrondo C, Flax J, Kucherov V , et al. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes[J]. PLoS One, 2016,11(1):e0147236. |
| [17] | Huang HW, Xie H, Ma X , et al. Upregulation of lncRNA PANDAR predicts poor prognosis and promotes cell proliferation in cervical cancer[J]. Eur Rev Med Pharmacol Sci, 2017,21(20):4529-4535. |
| [18] | Cao X, Xu J, Yue D . LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer[J]. Cancer Gene Ther, 2018,25(1/2):10-17. |
| [19] | Tuo Z, Zhang J, Xue W . LncRNA TP73-AS1 predicts the prognosis of bladder cancer patients and functions as a suppressor for bladder cancer by EMT pathway[J]. Biochem Biophys Res Commun, 2018,499(4):875-881. |
| [20] | Li HJ, Sun XM, Li ZK , et al. LncRNA UCA1promotes mitochondrial function of bladder cancer via the miR-195/ARL2 signaling pathway[J]. Cell Physiol Biochem, 2017,43(6):2548-2561. |
| [1] | 何丽杰,张春艳,王静. NEAT1、miR-27a-3p在阿尔茨海默病患者血清和脑脊液中的表达关系[J]. 北京大学学报(医学版), 2024, 56(2): 207-212. |
| [2] | 刘欢锐,彭祥,李森林,苟欣. 基于HER-2相关基因构建风险模型用于膀胱癌生存预后评估[J]. 北京大学学报(医学版), 2023, 55(5): 793-801. |
| [3] | 王飞,秦彩朋,杜依青,刘士军,李清,徐涛. 中危非肌层浸润性膀胱癌的最佳膀胱镜监测强度[J]. 北京大学学报(医学版), 2022, 54(4): 669-673. |
| [4] | 帅婷,刘娟,郭艳艳,金婵媛. 敲减长链非编码RNA MIR4697HG抑制骨髓间充质干细胞成脂向分化[J]. 北京大学学报(医学版), 2022, 54(2): 320-326. |
| [5] | 覃鸿泉,郑幽,王嫚娜,张峥嵘,牛祖彪,马骊,孙强,黄红艳,王小宁. 免疫相关GTP结合蛋白2的亚细胞定位分析[J]. 北京大学学报(医学版), 2020, 52(2): 221-226. |
| [6] | 黄海文,闫兵,尚美霞,刘漓波,郝瀚,席志军. 女性膀胱癌患者腹腔镜膀胱全切术与开放膀胱全切术的倾向性评分匹配比较[J]. 北京大学学报(医学版), 2019, 51(4): 698-705. |
| [7] | 覃子健,毕海,马潞林,黄毅,张帆. 肠代膀胱内原发肠源性腺癌1例[J]. 北京大学学报(医学版), 2018, 50(4): 737-739. |
| [8] | 唐兴国,颜野,邱敏,卢剑,陆敏,侯小飞,黄毅,马潞林. 单中心16年青年膀胱尿路上皮癌患者的诊治[J]. 北京大学学报(医学版), 2018, 50(4): 630-633. |
| [9] | 叶海云,许清泉,黄晓波,马凯,王晓峰. 卡介苗膀胱灌注治疗致结核性前列腺脓肿1例[J]. 北京大学学报(医学版), 2015, 47(6): 1039-1041. |
| [10] | 沈棋, 胡帅, 李峻, 王静华, 何群. 膀胱前列腺切除术中前列腺偶发癌发生率及临床病理特点分析[J]. 北京大学学报(医学版), 2014, 46(4): 515-518. |
| [11] | 景霞, 张其鹏, 国强华, 卢铭, 朱晓华, 石磊, 芮伟, 尚彤. 网上免费医学生物学数据库指南的建立[J]. 北京大学学报(医学版), 2004, 36(3): 322-326. |
| [12] | 马大龙. 计算机分析SARS病毒可能含有Caveolin结合区[J]. 北京大学学报(医学版), 2003, 35(z1): 139-139. |
| [13] | 芮伟, 张其鹏, 石磊, 卢铭, 景霞, 国强华, 尚彤. SARS冠状病毒RNA聚合酶编码区分析[J]. 北京大学学报(医学版), 2003, 35(z1): 137-138. |
| [14] | 芮伟, 张其鹏, 石磊, 卢铭, 景霞, 国强华, 尚彤. SARS冠状病毒可能编码蛋白质的三级结构预测[J]. 北京大学学报(医学版), 2003, 35(z1): 135-136. |
| [15] | 张其鹏, 石磊, 芮伟, 卢铭, 国强华, 景霞, 尚彤. SARS冠状病毒全基因组突变初步分析[J]. 北京大学学报(医学版), 2003, 35(z1): 130-131. |
|
||