北京大学学报(医学版) ›› 2019, Vol. 51 ›› Issue (3): 556-563. doi: 10.19723/j.issn.1671-167X.2019.03.027
中国唇腭裂患者Sonic hedgehog信号通路相关单核苷酸多态性的分析
张杰铌1,*,宋凤岐1,*,周绍楠1,郑晖1,彭丽颖1,张倩2,赵望泓3,张韬文4,李巍然1,周治波5,林久祥1△( ),陈峰2△(
),陈峰2△( )
)
- 1. 北京大学口腔医学院·口腔医院,正畸科 国家口腔疾病临床医学研究中心 口腔数字化医疗技术和材料国家工程实验室 口腔数字医学北京市重点实验室, 北京 100081
2. 北京大学口腔医学院·口腔医院中心实验室,北京 100081
3. 南方医科大学南方医院口腔科,广州 510000
4. 烟台市口腔医院口腔正畸科,山东烟台 264000
5. 北京大学口腔医学院·口腔医院口腔颌面外科,北京 100081
Analysis of single-nucleotide polymorphism of Sonic hedgehog signaling pathway in non-syndromic cleft lip and/or palate in the Chinese population
Jie-ni ZHANG1,*,Feng-qi SONG1,*,Shao-nan ZHOU1,Hui ZHENG1,Li-ying PENG1,Qian ZHANG2,Wang-hong ZHAO3,Tao-wen ZHANG4,Wei-ran LI1,Zhi-bo ZHOU5,Jiu-xiang LIN1△( ),Feng CHEN2△(
),Feng CHEN2△( )
)
- 1. Department of Orthodontics, Peking University School and Hospital of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Laboratory for Digital and Material Technology of Stomatology & Beijing Key Laboratory of Digital Stomatology, Beijing 100081, China
2. Department of Center Laboratory, Peking University School and Hospital of Stomatology, Beijing 100081, China
3. Department of Stomatology, Nanfang Hospital, Southern Medical University, Guangzhou 510000, China
4. Department of Orthodontics, Yantai Stomatological Hospital, Yantai 264000, Shandong, China
5. Department of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology, Beijing 100081, China
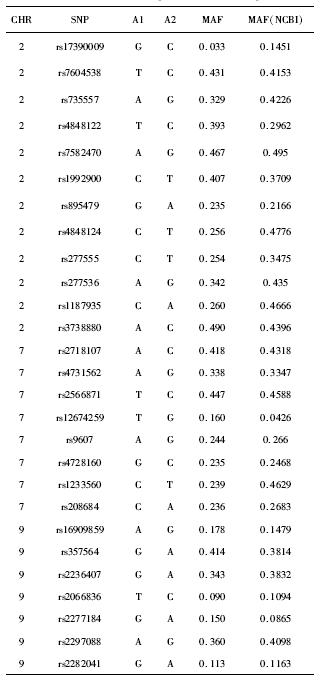
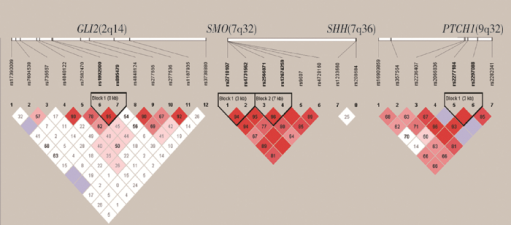
摘要: 目的 探讨Sonic hedgehog(Shh)信号通路相关的单核苷酸多态性(single-nucleotide polymorphism, SNP)与非综合征型唇腭裂(non-syndromic cleft lip and/or palate, NSCL/P) 之间的关联,并对唇腭裂疾病的致病风险因素进行探索。方法 收集197例个体的外周血(NSCL/P患者100例,健康对照97例),基于国际人类基因组单体型图计划中中国北京汉族人口数据,使用Haploview软件进行单倍体型分析和标签SNP选择。针对Shh 信号通路中的4个候选基因SHH、PTCH1、SMO和GLI2共选择了27个SNP。使用Sequenom质谱技术检测27个SNP在4个Shh信号通路中候选基因的基因型,并进行分析。结果 所选择的SNP基本涵盖了候选基因的潜在功能性SNP,其最小等位基因频率(minor allele frequency,MAF)>0.05:GLI2 73.5%, PTCH1 91.0%, SMO 100.0%, SHH 75.0%。发现位于SMO基因的SNP(rs12674259)和位于PTCH1基因的SNP(rs2066836)的基因型频率在NSCL/P病例组和对照组之间的差异有统计学意义。同时在4个候选基因所在的3条染色体(第2、7、9号染色体)中均发现了连锁不平衡,但在连锁不平衡单倍体型分析中,病例组和对照组之间的差异无统计学意义。结论 提示Shh信号通路参与NSCL/P的发生,其信号通路中关键基因的某些特殊SNP位点与唇腭裂相关,为NSCL/P的病因研究提供了新的探索方向,可能为NSCL/P的早期筛查与风险预测提供帮助。
中图分类号:
- R393
| [1] |
Mossey PA, Little J, Munger RG , et al. Cleft lip and palate[J]. Lancet, 2009,374(9703):1773-1785.
doi: 10.1016/S0140-6736(09)60695-4 |
| [2] | Dixon MJ, Marazita ML, Beaty TH , et al. Cleft lip and palate: understanding genetic and environmental influences[J]. Nat Rev Genet, 2011,12(3):167-178. |
| [3] |
Lidral AC, Murray JC . Genetic approaches to identify disease genes for birth defects with cleft lip/palate as a model[J]. Birth Defects Res B, 2004,70(12):893-901.
doi: 10.1002/(ISSN)1542-0760 |
| [4] | Schliekelman P, Slatkin M . Multiplex relative risk and estimation of the number of loci underlying an inherited disease[J]. Am J Hum Genet, 2003,71(6):1369-1385. |
| [5] |
Ludwig KU, Mangold E, Herms S , et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci[J]. Nat Genet, 2012,44(9):968-971.
doi: 10.1038/ng.2360 |
| [6] |
Marazita ML, Mooney MP . Current concepts in the embryology and genetics of cleft lip and cleft palate[J]. Clin Plast Surg, 2004,31(2):125-140.
doi: 10.1016/S0094-1298(03)00138-X |
| [7] |
Zhang J, Zhou S, Zhang Q , et al. Proteomic analysis of RBP4/vitamin A in children with cleft lip and/or palate[J]. J Dent Res, 2014,93(6):547-552.
doi: 10.1177/0022034514530397 |
| [8] | Hu D, Helms JA . The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis[J]. Development, 1999,126(21):4873-4884. |
| [9] |
Vieira AR, Castilla EE, Ming JE , et al. Mutational analysis of the Sonic hedgehog gene in 220 newborns with oral clefts in a South American (ECLAMC) population[J]. Am J Med Genet, 2002,108(1):12-15.
doi: 10.1002/ajmg.10204 |
| [10] |
Han J, Mayo J, Xu X , et al. Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice[J]. Development, 2009,136(24):4225-4233.
doi: 10.1242/dev.036723 |
| [11] | Shimamura K, Hartigan DJ, Martinez S , et al. Longitudinal organization of the anterior neural plate and neural tube[J]. Development, 1995,121(12):3923-3933. |
| [12] |
Belloni E, Muenke M, Roessler E , et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly[J]. Nat Genet, 1996,14(3):353-356.
doi: 10.1038/ng1196-353 |
| [13] |
Chiang C, Litingtung Y, Lee E , et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function[J]. Nature, 1996,383(6599):407-413.
doi: 10.1038/383407a0 |
| [14] |
Rahnama F, Shimokawa T, Lauth M , et al. Inhibition of GLI1 gene activation by Patched1[J]. Biochem J, 2006,394(Pt 1):19-26.
doi: 10.1042/BJ20050941 |
| [15] |
Beaty TH, Fallin MD, Hetmanski JB , et al. Haplotype diversity in 11 candidate genes across four populations[J]. Genetics, 2005,171(1):259-267.
doi: 10.1534/genetics.105.043075 |
| [16] |
Xing J, Witherspoon DJ, Watkins WS , et al. HapMap tag SNP transferability in multiple populations: general guidelines[J]. Genomics, 2008,92(1):41-51.
doi: 10.1016/j.ygeno.2008.03.011 |
| [17] |
Jiang L, Zhang C, Li Y , et al. A non-synonymous polymorphism Thr115Met in the EpCAM gene is associated with an increased risk of breast cancer in Chinese population[J]. Breast Cancer Res Treat, 2011,126(2):487-495.
doi: 10.1007/s10549-010-1094-6 |
| [18] |
Ong KL, Li M, Tso AK , et al. Association of genetic variants in the adiponectin gene with adiponectin level and hypertension in Hong Kong Chinese[J]. Eur J Endocrinol, 2010,163(2):251-257.
doi: 10.1530/EJE-10-0251 |
| [19] |
Barrett JC, Fry B, Maller J , et al. Haploview: analysis and visualization of LD and haplotype maps[J]. Bioinformatics, 2005,21(2):263-265.
doi: 10.1093/bioinformatics/bth457 |
| [20] |
Farias LC, Gomes CC, Brito JA , et al. Loss of heterozygosity of the PTCH gene in ameloblastoma[J]. Hum Pathol, 2012,43(8):1229-1233.
doi: 10.1016/j.humpath.2011.08.026 |
| [21] |
Cohen MM . Holoprosencephaly: clinical, anatomic, and molecular dimensions.[J]. Birth Defects Res, 2006,76(9):658-673.
doi: 10.1002/(ISSN)1542-0760 |
| [22] | Carter TC, Molloy AM, Pangilinan F , et al. Testing reported associations of genetic risk factors for oral clefts in a large Irish study population[J]. Birth Defects Res, 2010,88(2):84-93. |
| [23] | Brand M, Heisenberg CP, Warga RM , et al. Mutations affecting development of the midline and general body shape during zebrafish embryogenesis[J]. Development, 1996,123(6):129-142. |
| [24] |
Eberhart JK, Swartz ME, Crump JG , et al. Early hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development[J]. Development, 2006,133(6):1069-1077.
doi: 10.1242/dev.02281 |
| [25] | Yahya MJ, Ismail PB, Nordin NB , et al. CNDP1, NOS3, and MnSOD polymorphisms as risk factors for diabetic nephropathy among type 2 diabetic patients in Malaysia[J]. J Nutr Metab, 2019(3):1-13. |
| [26] | Zhuo M, Zhuang X, Tang W , et al. Theimpact of IL-16 3’UTR polymorphism rs859 on lung carcinoma susceptibility among Chinese han individuals[J]. Biomed Res Int, 2018,12(24):1-10. |
| [27] |
Beaty TH, Hetmanski JB, Fallin MD , et al. Analysis of candidate genes on chromosome 2 in oral cleft case-parent trios from three populations[J]. Hum Genet, 2006,120(4):501-518.
doi: 10.1007/s00439-006-0235-9 |
| [28] |
Levi B, James AW, Nelson ER , et al. Role of Indian hedgehog signaling in palatal osteogenesis[J]. Plast Reconstr Surg, 2011,127(3):1182-1190.
doi: 10.1097/PRS.0b013e3182043a07 |
| [29] |
Helms JA, Kim CH, Hu D , et al. Sonic hedgehog participates in craniofacial morphogenesis and is down-regulated by teratogenic doses of retinoic acid[J]. Dev Biol, 1997,187(1):1-35.
doi: 10.1006/dbio.1997.8572 |
| [1] | 薛恩慈, 陈曦, 王雪珩, 王斯悦, 王梦莹, 李劲, 秦雪英, 武轶群, 李楠, 李静, 周治波, 朱洪平, 吴涛, 陈大方, 胡永华. 中国人群非综合征型唇裂伴或不伴腭裂的单核苷酸多态性遗传度[J]. 北京大学学报(医学版), 2024, 56(5): 775-780. |
| [2] | 侯天姣,周治波,王竹青,王梦莹,王斯悦,彭和香,郭煌达,李奕昕,章涵宇,秦雪英,武轶群,郑鸿尘,李静,吴涛,朱洪平. 转化生长因子β信号通路与非综合征型唇腭裂发病风险的基因-基因及基因-环境交互作用[J]. 北京大学学报(医学版), 2024, 56(3): 384-389. |
| [3] | 王梦莹,李文咏,周仁,王斯悦,刘冬静,郑鸿尘,周治波,朱洪平,吴涛,胡永华. WNT信号通路基因位点单体型与中国汉族人群非综合征型唇腭裂发病风险的关联[J]. 北京大学学报(医学版), 2022, 54(3): 394-399. |
| [4] | 刘建,王宪娥,吕达,乔敏,张立,孟焕新,徐莉,毛铭馨. 广泛型侵袭性牙周炎患者牙根形态异常与相关致病基因的关联[J]. 北京大学学报(医学版), 2021, 53(1): 16-23. |
| [5] | 王梦莹,李文咏,周仁,王斯悦,刘冬静,郑鸿尘,李静,李楠,周治波,朱洪平,吴涛,胡永华. WNT代谢通路相关基因与中国人群非综合征型唇腭裂发病风险的交互作用[J]. 北京大学学报(医学版), 2020, 52(5): 815-820. |
| [6] | 李文咏,王梦莹,周仁,王斯悦,郑鸿尘,朱洪平,周治波,吴涛,王红,石冰. 中国人群Hedgehog通路基因与非综合征型唇腭裂的亲源效应[J]. 北京大学学报(医学版), 2020, 52(5): 809-814. |
| [7] | 田艳,朱军. p53 rs1625895基因多态性与弥漫大B细胞淋巴瘤预后相关性分析[J]. 北京大学学报(医学版), 2019, 51(5): 791-796. |
| [8] | 周仁,郑鸿尘,李文咏,王梦莹,王斯悦,李楠,李静,周治波,吴涛,朱洪平. 利用二代测序数据探索SPRY基因家族与中国人群非综合征型唇腭裂的关联[J]. 北京大学学报(医学版), 2019, 51(3): 564-570. |
| [9] | 王丹丹,甘业华,马绪臣,孟娟红. ADAMTS14基因单核苷酸多态性与汉族女性颞下颌关节骨关节炎的相关性研究[J]. 北京大学学报(医学版), 2018, 50(2): 279-283. |
| [10] | 周彦秋, 林久祥, 张波. sonic hedgehog及patched基因在角化囊肿中的表达[J]. 北京大学学报(医学版), 2002, 34(4): 380-383. |
| [11] | 周彦秋, 林久祥. RT-PCR法研究sonic hedgehog基因及其受体patched在小鼠牙胚钟状晚期的表达[J]. 北京大学学报(医学版), 2002, 34(2): 137-139. |
|
||