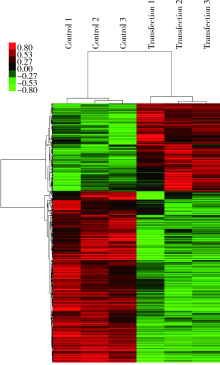
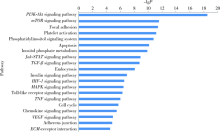
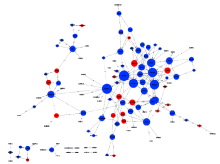
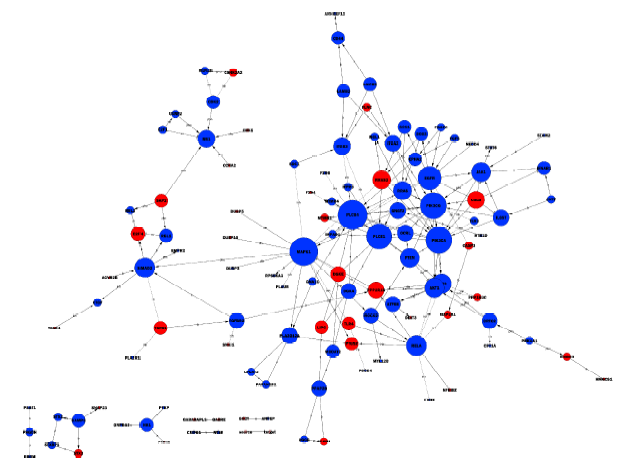
Journal of Peking University(Health Sciences) ›› 2019, Vol. 51 ›› Issue (2): 221-227. doi: 10.19723/j.issn.1671-167X.2019.02.004
Previous Articles Next Articles
Role of miR-106b-5p in the regulation of gene profiles in endothelial cells
Jing ZHANG,Su-fang LI,Hong CHEN( ),Jun-xian SONG
),Jun-xian SONG
- Center for Cardiovascular Translational Research, Peking University People’s Hospital, Beijing 100044, China;
CLC Number:
- R541.4
| [1] |
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function[J]. Cell, 2004,116(2):281-297.
doi: 10.1016/S0092-8674(04)00045-5 |
| [2] |
Small EM, Olson EN . Pervasive roles of microRNAs in cardiovascular biology[J]. Nature, 2011,469(7330):336-342.
doi: 10.1038/nature09783 |
| [3] |
Ren J, Zhang J, Xu N , et al. Signature of circulating microRNAs as potential biomarkers in vulnerable coronary artery disease[J]. PLoS One, 2013,8(12):e80738.
doi: 10.1371/journal.pone.0080738 |
| [4] |
Li P, Shen M, Gao F , et al. An antagomir to microRNA-106b-5p ameliorates cerebral ischemia and reperfusion injury in rats via inhibiting apoptosis and oxidative stress[J]. Mol Neurobiol, 2017,54(4):2901-2921.
doi: 10.1007/s12035-016-9842-1 |
| [5] |
Li N, Miao Y, Shan Y , et al. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer[J]. Cell Death Dis, 2017,8(5):e2796.
doi: 10.1038/cddis.2017.119 |
| [6] |
Wang J, Haubrock M, Cao KM , et al. Regulatory coordination of clustered microRNAs based on microRNA-transcription factor regulatory network[J]. BMC Syst Biol, 2011,5(12):199.
doi: 10.1186/1752-0509-5-199 |
| [7] |
Prieto C, Risueño A, Fontanillo C , et al. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles[J]. PLoS One, 2008,3(12):e3911.
doi: 10.1371/journal.pone.0003911 |
| [8] |
Zhang J, Li SF, Chen H , et al. MiR-106b-5p inhibits tumor necrosis factor-α-induced apoptosis by targeting phosphatase and tensin homolog deleted on chromosome 10 in vascular endothelial cells[J]. Chin Med J (Engl), 2016,129(12):1406-1412.
doi: 10.4103/0366-6999.183414 |
| [9] |
Ogata H, Goto S, Sato K , et al. KEGG: Kyoto Encyclopedia of Genes and Genomes[J]. Nucleic Acids Res, 1999,27(1):29-34.
doi: 10.1093/nar/27.1.29 |
| [10] |
Du J, Yuan Z, Ma Z , et al. KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model[J]. Mol Biosyst, 2014,10(9):2441-2447.
doi: 10.1039/C4MB00287C |
| [11] |
Kanehisa M, Goto S, Sato Y , et al. KEGG for integration and interpretation of large-scale molecular data sets[J]. Nucleic Acids Res, 2012,40(Database issue):D109-114.
doi: 10.1093/nar/gkr988 |
| [12] |
Ross R . Atherosclerosis: an inflammatory disease[J]. N Engl J Med, 1999,340(2):115-126.
doi: 10.1056/NEJM199901143400207 |
| [13] | 任景怡, 许宁, 韩冠平 , 等. microRNAs 参与动脉粥样硬化疾病的发生[J]. 中国生物化学与分子生物学报, 2011,27(6):511-515. |
| [14] |
Zampetaki A, Willeit P, Drozdov I , et al. Profiling of circulating microRNAs: from single biomarkers to re-wired networks[J]. Cardiovasc Res, 2012,93(4):555-562.
doi: 10.1093/cvr/cvr266 |
| [15] |
Weber C, Noels H . Atherosclerosis: current pathogenesis and therapeutic options[J]. Nat Med, 2011,17(11):1410-1422.
doi: 10.1038/nm.2538 |
| [16] |
Zernecke A, Weber C . Chemokines in the vascular inflammatory response of atherosclerosis[J]. Cardiovasc Res, 2010,86(2):192-201.
doi: 10.1093/cvr/cvp391 |
| [17] |
Lutgens E, Daemen MJ . Transforming growth factor-β: a local or systemic mediator of plaque stability?[J]. Circ Res, 2001,89(10):853-855.
doi: 10.1161/res.89.10.853 |
| [18] |
Lutgens E, Gijbels M, Smook M , et al. Transforming growth factor-beta mediates balance between inflammation and fibrosis during plaque progression[J]. Arterioscler Thromb Vasc Biol, 2002,22(6):975-982.
doi: 10.1161/01.ATV.0000019729.39500.2F |
| [1] | Jing HE,Zhongze FANG,Ying YANG,Jing LIU,Wenyao MA,Yong HUO,Wei GAO,Yangfeng WU,Gaoqiang XIE. Relationship between lipid metabolism molecules in plasma and carotid atheroscle-rotic plaques, traditional cardiovascular risk factors, and dietary factors [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 722-728. |
| [2] | Xiaomeng REN,Kaiyi LI,Chunlei LI. Detection of molecular affecting sensitivity to local glucocorticoid therapy in oral lichen planus through transcriptome sequencing [J]. Journal of Peking University (Health Sciences), 2024, 56(1): 32-38. |
| [3] | Wen-gen LI,Xiao-dong GU,Rui-qiang WENG,Su-dong LIU,Chao CHEN. Expression and clinical significance of plasma exosomal miR-34-5p and miR-142-3p in systemic sclerosis [J]. Journal of Peking University (Health Sciences), 2023, 55(6): 1022-1027. |
| [4] | HUANG Li-dong,GONG Wei-yu,DONG Yan-mei. Effects of bioactive glass on proliferation, differentiation and angiogenesis of human umbilical vein endothelial cells [J]. Journal of Peking University (Health Sciences), 2021, 53(2): 371-377. |
| [5] | Teng-fei LIU,Tao LIN,Li-hui REN,Guang-ping LI,Jian-jun PENG. Association between CMTM5 gene and coronary artery disease and the relative mechanism [J]. Journal of Peking University (Health Sciences), 2020, 52(6): 1082-1087. |
| [6] | Yan XUAN,Yu CAI,Xiao-xuan WANG,Qiao SHI,Li-xin QIU,Qing-xian LUAN. Effect of Porphyromonas gingivalis infection on atherosclerosis in apolipoprotein-E knockout mice [J]. Journal of Peking University (Health Sciences), 2020, 52(4): 743-749. |
| [7] | Xiao WANG,Dan HE,Wen-ting LI,SIYITI Adila·,Rui HAN,Ying DONG. Characteristic and clinical significance of microRNA expression between 144 Uygur and Han women with endometrial carcinoma [J]. Journal of Peking University (Health Sciences), 2020, 52(3): 570-577. |
| [8] | Jing XIE,Yu-ming ZHAO,Nan-quan RAO,Xiao-tong WANG,Teng-jiao-zi FANG,Xiao-xia LI,Yue ZHAI,Jing-zhi LI,Li-hong GE,Yuan-yuan WANG. Comparative study of differentiation potential of mesenchymal stem cells derived from orofacial system into vascular endothelial cells [J]. Journal of Peking University(Health Sciences), 2019, 51(5): 900-906. |
| [9] | Wei ZHANG,Ying-jiang YE,Xian-wen REN,Jing HUANG,Zhan-long SHEN. Detection of preoperative chemoradiotherapy sensitivity molecular characteristics of rectal cancer by transcriptome second generation sequencing [J]. Journal of Peking University(Health Sciences), 2019, 51(3): 542-547. |
| [10] | LIU Ying-jun, OUYANG Xiang-ying, WANG Yu-guang, LYU Pei-jun, AN Na. Role of vitamin K-dependent protein Gas6 in the expression of endothelial cell adhesion molecule-1 and chemokines induced by Porphyromonas gingivalis lipopolysaccharide [J]. Journal of Peking University(Health Sciences), 2018, 50(1): 20-25. |
| [11] | CAI Yi, GUO Hao, LI Han-zhong, WANG Wen-da, ZHANG Yu-shi. MicroRNA differential expression profile in tuberous sclerosis complex cell line TSC2-/- MEFs and normal cell line TSC2+/+ MEFs [J]. Journal of Peking University(Health Sciences), 2017, 49(4): 580-584. |
| [12] | XIAO Tian-yi, LIU Yan, LI Ji-lai, WANG Rui-tong, DU Ji-chen. Diagnostic value of carotid atherosclerosis score for ischemic stroke [J]. Journal of Peking University(Health Sciences), 2016, 48(6): 1000-1005. |
| [13] | HAN Jin-tao, LI Xuan, HE Qing-yuan, ZHAO Hai-yan, YE Shan, DONG Guo-xiang, LUAN Jing-yuan, WANG Chang-ming. Endovascular treatment in cerebral artery tandem lesions [J]. Journal of Peking University(Health Sciences), 2016, 48(1): 149-153. |
| [14] | JIA Ling-fei, GAN Ye-hua, YU Guang-yan. Relationships between microRNA expressions and prognosis in patients with tongue squamous cell carcinoma and the mechanisms microRNA regulating tongue squamous cell carcinoma biological behavior [J]. Journal of Peking University(Health Sciences), 2016, 48(1): 5-9. |
| [15] | LI Peng, WAN Meng, LIU Jian-ru, LI Liang-zhong, ZHANG Da-kun. Effect of peroxisome proliferator-activated receptor-γ on endothelial cells oxidative stress induced by Porphyromonas gingivalis [J]. Journal of Peking University(Health Sciences), 2015, 47(6): 977-982. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 329
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 1007
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
Cited |
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| Shared | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Discussed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
||