北京大学学报(医学版) ›› 2021, Vol. 53 ›› Issue (3): 602-607. doi: 10.19723/j.issn.1671-167X.2021.03.028
基于肿瘤基因组图谱数据库探索性筛选潜在泛癌生物标志物
周川,马雪,邢云昆,李璐迪,陈洁,姚碧云,傅娟玲,赵鹏Δ( )
)
- 北京大学公共卫生学院毒理学系,食品安全毒理学研究与评价北京市重点实验室,北京 100191
Exploratory screening of potential pan-cancer biomarkers based on The Cancer Genome Atlas database
ZHOU Chuan,MA Xue,XING Yun-kun,LI Lu-di,CHEN Jie,YAO Bi-yun,FU Juan-ling,ZHAO PengΔ( )
)
- Department of Toxicology, Beijing Key Laboratory of Toxicological Research and Risk Assessment for Food Safety, Peking University School of Public Health, Beijing 100191, China
摘要:
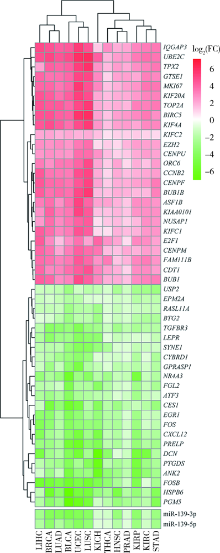
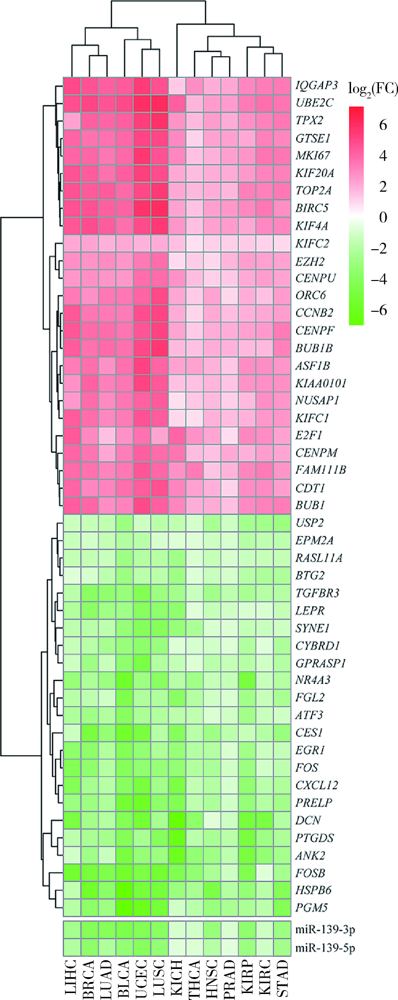
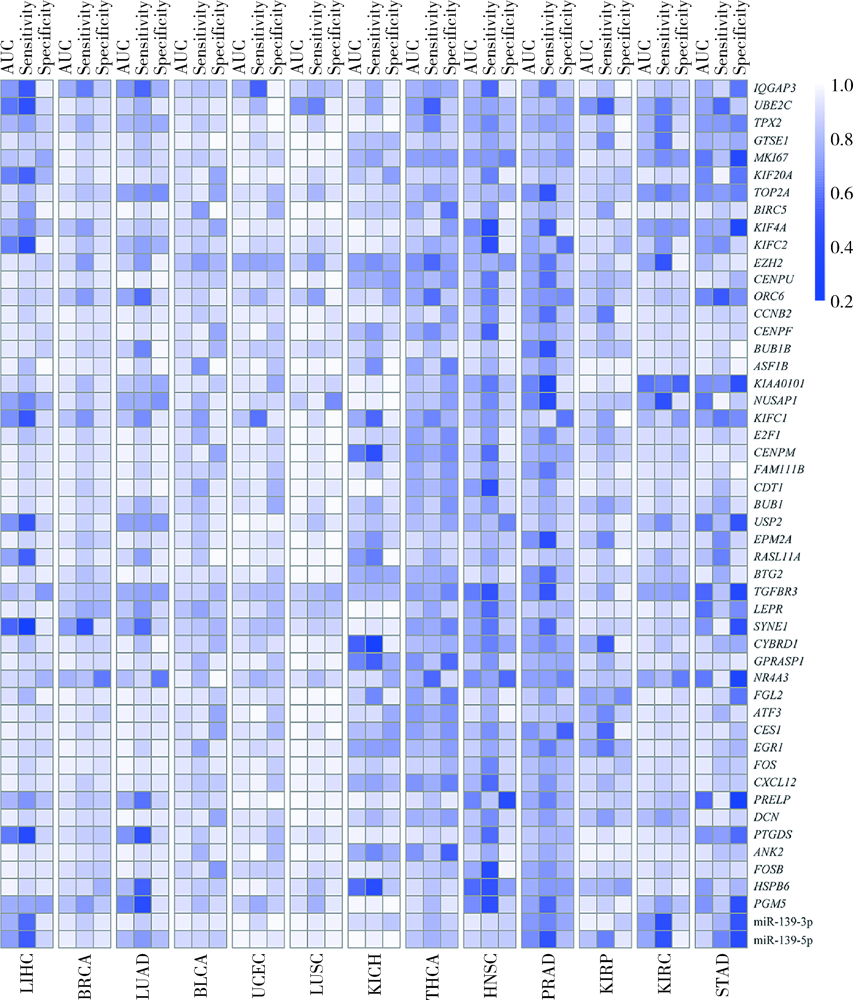
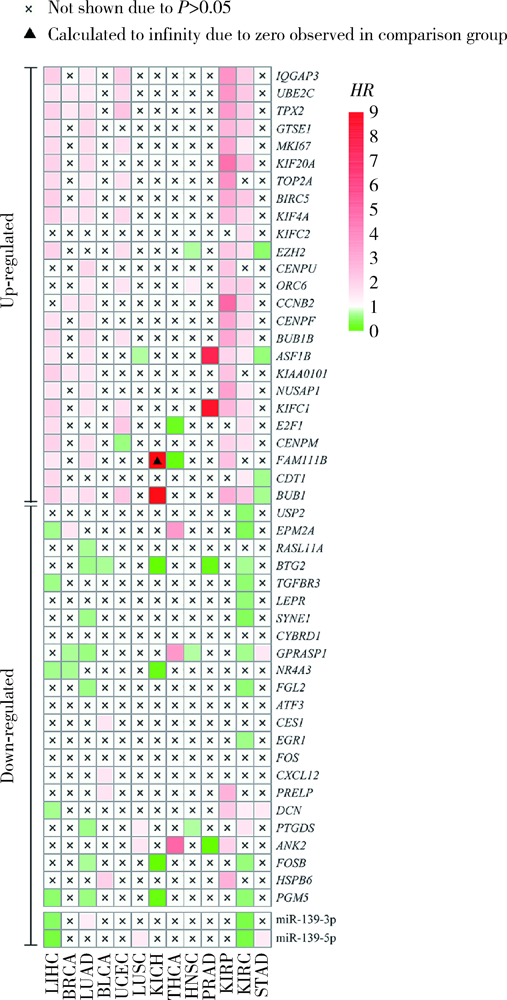
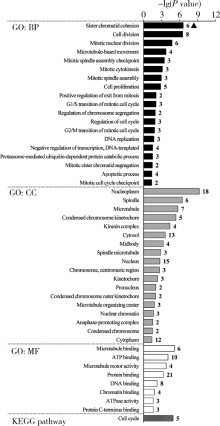
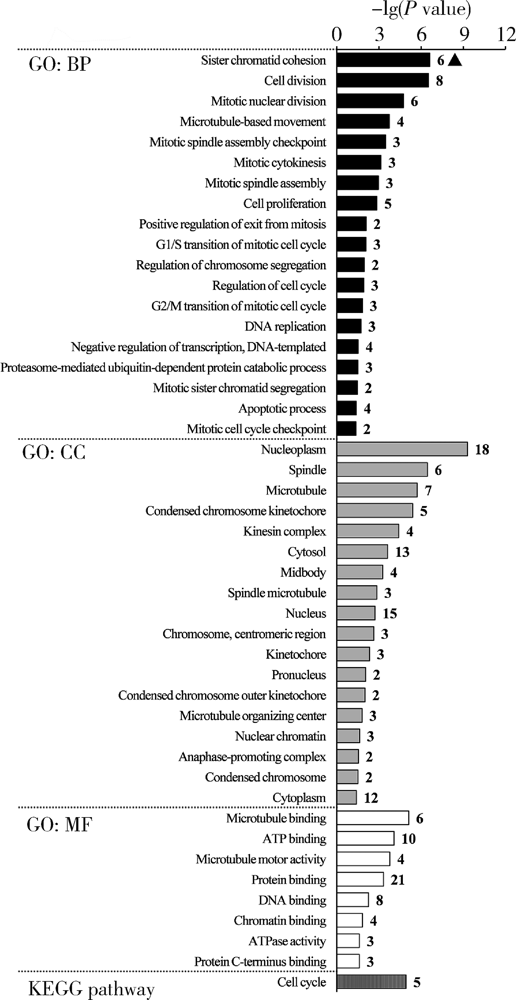
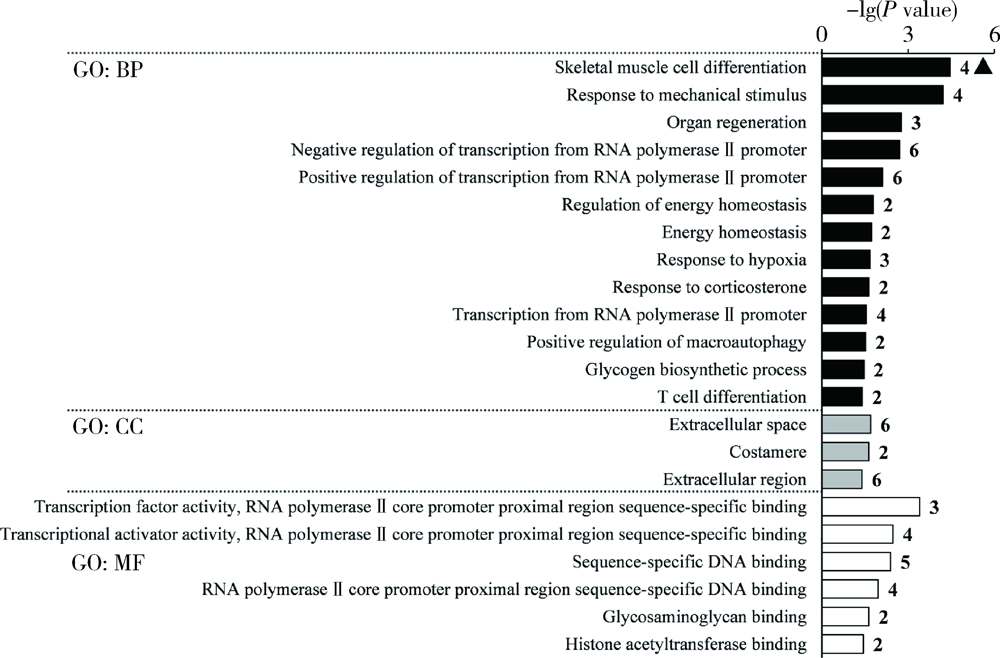
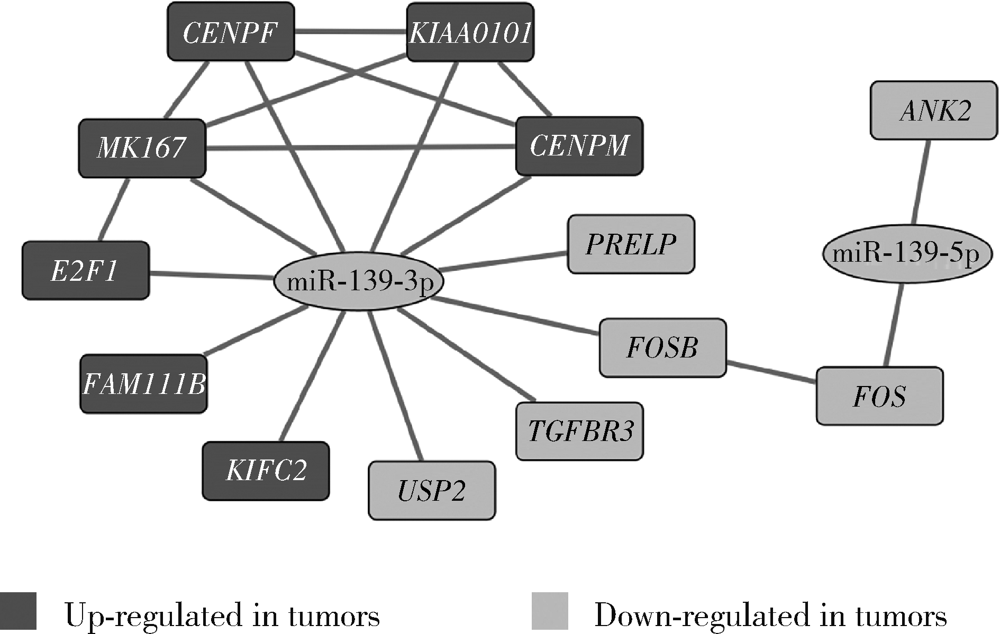
目的: 基于肿瘤基因组图谱(The Cancer Genome Atlas, TCGA)数据库筛选潜在泛癌生物标志物,为多种肿瘤的诊断和预后评估提供帮助。方法: 利用“GDC Data Transfer Tool”和“GDCRNATools”软件包获取TCGA数据库,完成数据整理,将13种肿瘤纳入研究。以错误发现率(false discovery rate, FDR) <0.05且差异倍数(fold change, FC) >1.5作为差异表达标准,筛选在13种肿瘤中均上调或均下调的基因和微小RNA(microRNAs,miRNAs)。利用受试者工作特征曲线(receiver operating characteristic curve, ROC曲线)的曲线下面积(area under the curve, AUC)、最佳截断值及对应的灵敏度和特异度反映诊断价值。利用Kaplan-Meier法计算生存概率后进行对数秩(log-rank)检验并计算风险比(hazard ratio, HR)反映预后评估价值。利用DAVID工具对差异表达基因进行GO (Gene Ontology)、KEGG (Kyoto Encyclopedia of Genes and Genomes)富集分析。利用STRING和TargetScan工具对差异表达基因和miRNAs进行调控网络分析。结果: 共发现48个基因和2个miRNAs在13种肿瘤中均差异表达,其中25个基因均表达上调,23个基因和2个miRNAs均表达下调。多数差异表达基因和miRNAs区分病例和对照的能力较好,AUC、灵敏度和特异度可达0.8~0.9。生存分析结果显示,差异表达基因和miRNAs与多种肿瘤患者的生存显著相关,且多数上调基因是患者生存的危险因素(HR>1),而多数下调基因是患者生存的保护因素(0<HR<1)。GO和KEGG富集分析显示,差异表达基因多富集于与细胞增殖有关的生物学事件。在调控网络分析中,共13个基因和2个miRNAs存在调控和相互作用关系。结论: 在13种肿瘤中均差异表达的48个基因和2个miRNAs可能作为潜在泛癌生物标志物,为多种肿瘤的诊断和预后评估提供帮助,并为发展肿瘤治疗的广谱靶点提供线索。
中图分类号:
- R730.43
| [1] |
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018,68(6):394-424.
doi: 10.3322/caac.v68.6 |
| [2] |
Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study[J]. Nat Rev Cancer, 2016,16(8):525-537.
doi: 10.1038/nrc.2016.56 |
| [3] | 王印祥. 泛肿瘤研究和肿瘤免疫研究: 未来抗肿瘤药的发展趋势[J]. 中国药物化学杂志, 2015,25(2):149-152. |
| [4] |
Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration[J]. Nat Genet, 2013,45(10):1134-1140.
doi: 10.1038/ng.2760 |
| [5] | 陈熹, 张峻峰, 曾科, 等. 血清microRNA: 一种非侵入性的肿瘤标志物[J]. 生命科学, 2010,22(7):649-654. |
| [6] |
Li RD, Qu H, Wang SB, et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC[J]. Bioinformatics, 2018,34(14):2515-2517.
doi: 10.1093/bioinformatics/bty124 |
| [7] |
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources[J]. Nat Protoc, 2009,4(1):44-57.
doi: 10.1038/nprot.2008.211 pmid: 19131956 |
| [8] |
Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets[J]. Nucleic Acids Res, 2019,47(D1):D607-D613.
doi: 10.1093/nar/gky1131 |
| [9] |
Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs[J]. Elife, 2015,4:e05005.
doi: 10.7554/eLife.05005 |
| [10] |
Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project[J]. Nat Genet, 2013,45(10):1113-1120.
doi: 10.1038/ng.2764 |
| [11] |
Penault-Llorca F, Radosevic-Robin N. Ki67 assessment in breast cancer: an update[J]. Pathology, 2017,49(2):166-171.
doi: S0031-3025(16)40573-8 pmid: 28065411 |
| [12] |
Yuniati L, Scheijen B, van der Meer LT, et al. Tumor suppressors BTG1 and BTG2: Beyond growth control[J]. J Cell Physiol, 2019,234(5):5379-5389.
doi: 10.1002/jcp.27407 |
| [13] |
Bhattacharjee S, Rajaraman P, Jacobs KB, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits[J]. Am J Hum Genet, 2012,90(5):821-835.
doi: 10.1016/j.ajhg.2012.03.015 |
| [14] |
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation[J]. Cell, 2011,144(5):646-674.
doi: 10.1016/j.cell.2011.02.013 |
| [15] |
Laine A, Westermarck J. Molecular pathways: harnessing E2F1 regulation for prosenescence therapy in p53-defective cancer cells[J]. Clin Cancer Res, 2014,20(14):3644-3650.
doi: 10.1158/1078-0432.CCR-13-1942 pmid: 24788101 |
| [16] |
Xiao YS, Najeeb RM, Ma D, et al. Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms[J]. J Exp Clin Cancer Res, 2019,38(1):458.
doi: 10.1186/s13046-019-1444-0 |
| [17] |
Sun JB, Huang JZ, Lan J, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer[J]. Cancer Cell Int, 2019,19(1):264.
doi: 10.1186/s12935-019-0986-8 |
| [1] | 郭倩,陈绪勇,苏茵. 白细胞介素2信号通路相关分子与系统性红斑狼疮[J]. 北京大学学报(医学版), 2016, 48(6): 1100-1104. |
| [2] | 张驰. 成骨细胞特异性转录因子Osterix对骨形成作用的分子机制[J]. 北京大学学报(医学版), 2012, 44(5): 659-665. |
| [3] | 吕平, 高学军. 釉质缺陷临床表现型分析对分子调控研究的启示[J]. 北京大学学报(医学版), 2009, 41(1): 121-123. |
| [4] | 田华, Carrie FANG, Paul E.DICESARE. 骨形态发生蛋白-2对软骨寡聚基质蛋白在软骨细胞内表达的影响[J]. 北京大学学报(医学版), 2003, 35(3): 317-320. |
| [5] | 孔灵玲, 方伟岗, 钟镐镐, 衡万杰, 李燕, 吴秉铨. 金属蛋白酶MMP-9可调控型表达与人黑色素瘤细胞侵袭表型的相关性[J]. 北京大学学报(医学版), 2003, 35(1): 7-11. |
| [6] | 石红霞, 李莉, 江滨, 卢锡京, 韩伟, 丘镜莹, 傅剑锋, 王德炳, 崔健英. 急性白血病患者bcl-xL, mdr-1,mrp 基因表达及临床意义[J]. 北京大学学报(医学版), 2002, 34(1): 88-90. |
|
||