北京大学学报(医学版) ›› 2022, Vol. 54 ›› Issue (1): 23-30. doi: 10.19723/j.issn.1671-167X.2022.01.005
计算机模拟亚甲基蓝与牙龈卟啉单胞菌部分蛋白的分子对接
袁临天1,2,马利沙2,刘润园3,齐伟1,2,张栌丹2,4,王贵燕2,5,王宇光2,△( )
)
- 1.北京大学口腔医学院·口腔医院 综合科,国家口腔疾病临床医学研究中心,口腔数字化医疗技术和材料国家工程实验室,口腔数字医学北京市重点实验室,北京 100081
2.北京大学口腔医学院·口腔医院 口腔医学数字化研究中心,北京 100081
3.大连医科大学口腔医学院牙体牙髓科,辽宁大连 116044
4.北京大学口腔医学院·口腔医院门诊部,北京 100081
5.北京大学口腔医学院·口腔医院 儿童口腔科,北京 100081
Computer simulation of molecular docking between methylene blue and some proteins of Porphyromonas gingivalis
YUAN Lin-tian1,2,MA Li-sha2,LIU Run-yuan3,QI wei1,2,ZHANG Lu-dan2,4,WANG Gui-yan2,5,WANG Yu-guang2,△( )
)
- 1. Department of General Medicine, Peking University School and Hospital of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Laboratory for Digital and Material Technology of Stomatology & Beijing Key Laboratory of Digital Stomatology, Beijing 100081, China
2. Center for Digital Dentistry, Peking University School and Hospital of Stomatology, Beijing 100081, China
3. Department of Endodontics, College of Stomatology, Dalian Medical University, Dalian 116044, Liaoning, China
4. First Clinical Division, Peking University School and Hospital of Stomatology, Beijing 100081, China
5. Department of Pediatric Dentistry, Peking University School and Hospital of Stomatology, Beijing 100081, China
摘要:
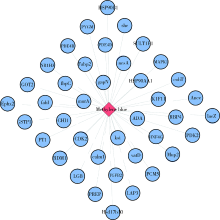
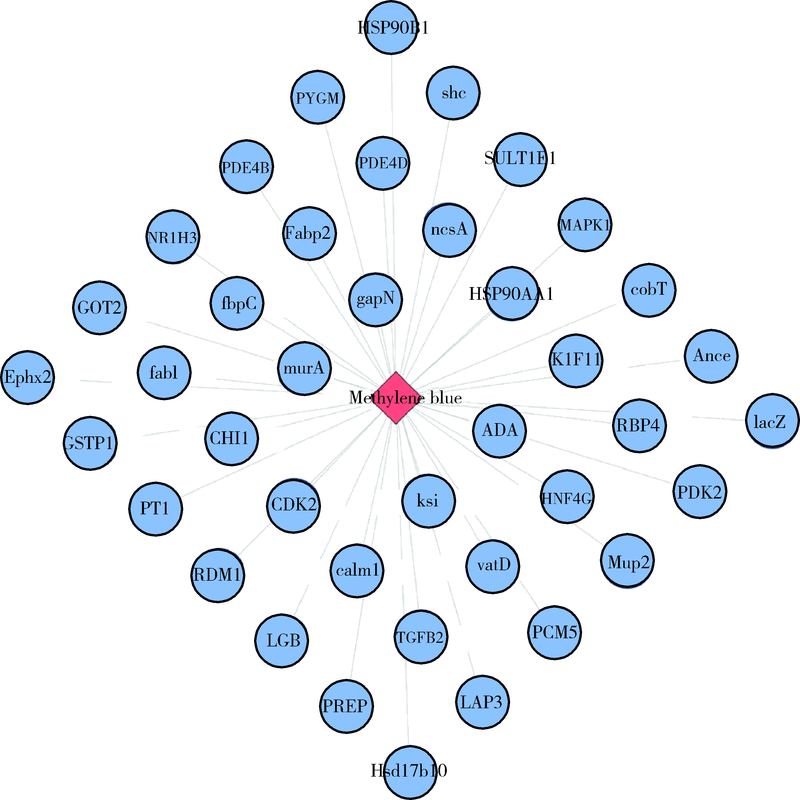
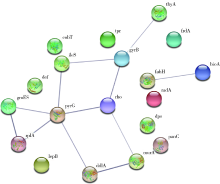
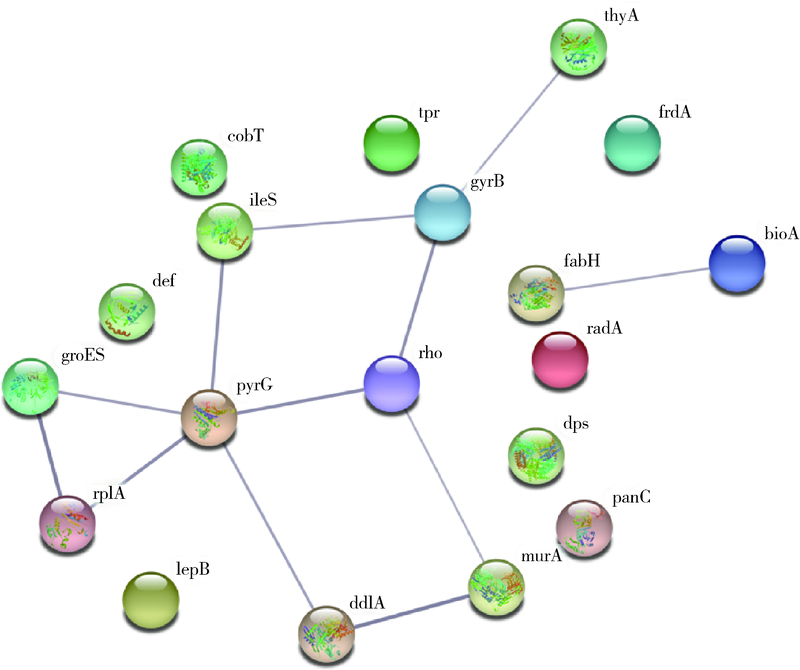
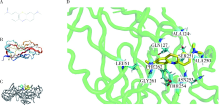
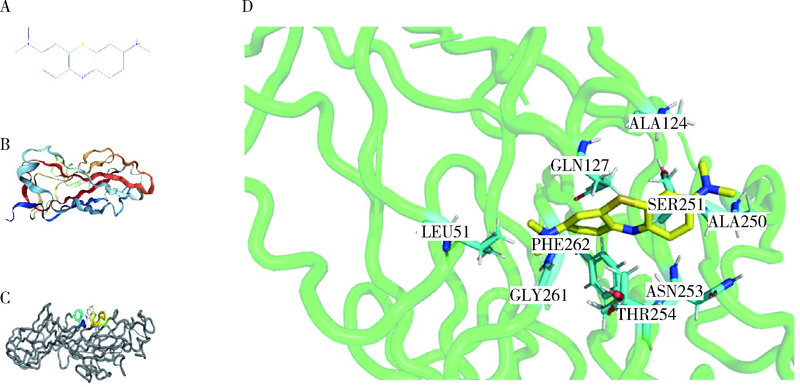
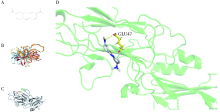
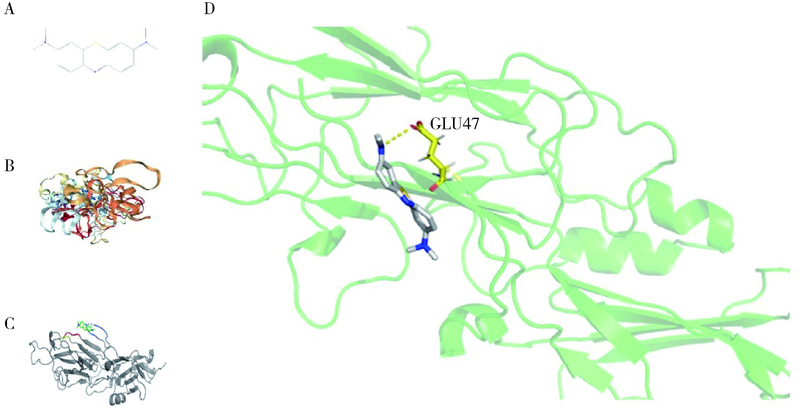
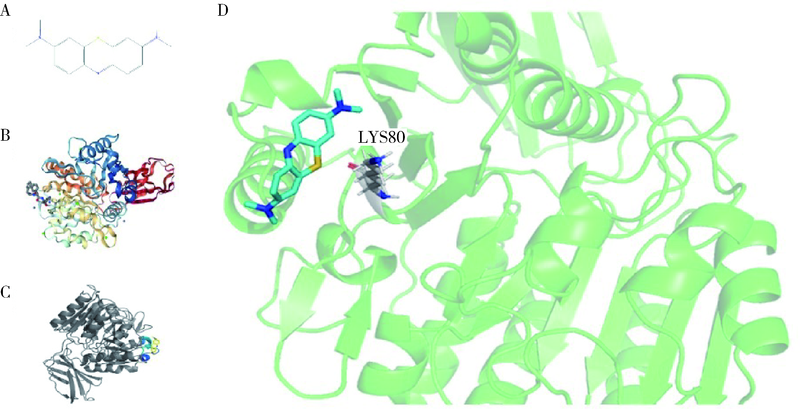
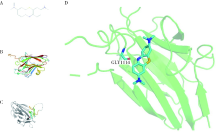
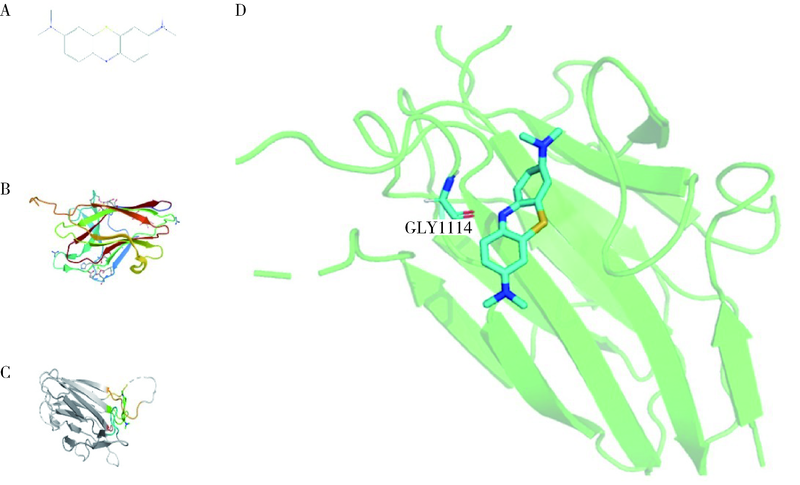
目的: 使用计算机模拟的靶点预测与分子对接的方法,研究抗菌光动力疗法中光敏剂与细菌结合的靶点,并计算结合能。方法: 在Uniprot数据库和RCSB PDB数据库中获取并汇总牙龈卟啉单胞菌(Porphyromonas gingivalis,Pg)的蛋白名称;在SciFinder数据库、PubChem数据库、ChemSpider数据库和Chemical Book中筛选并对比亚甲基蓝的结构图,并用ChemBioDraw软件绘制确认;在PharmMapper数据库对亚甲基蓝三维结构进行靶点预测,并用Cytoscape 软件构建可视化网络图;在 String 数据库中构建亚甲基蓝靶点与Pg蛋白交集的相互作用网络;选择FimA、Mfa4、RgpB、Kgp K1蛋白,使用AutoDock软件计算亚甲基蓝与上述蛋白的对接能量,并进行分子对接。结果: 靶点预测结果显示,268个亚甲基蓝潜在靶点和1 865个Pg的蛋白之间有19个共同的靶点,这19个靶点为:groS、radA、rplA、dps、fabH、pyrG、thyA、panC、RHO、frdA、ileS、bioA、def、ddl、TPR、murA、lepB、cobT、gyrB。分子对接结果显示,亚甲基蓝能与FimA蛋白的9个位点结合,结合能-6.26 kcal/mol;与Mfa4蛋白的4个位点和氢键形成位点GLU47结合,结合能-5.91 kcal/mol;与RgpB蛋白的氢键形成位点LYS80结合,结合能-5.14 kcal/mol;与Kgp K1蛋白的6个位点和氢键形成位点GLY1114结合,结合能-5.07 kcal/mol。结论: 计算机模拟的靶点预测与分子对接技术可以初步揭示亚甲基蓝与Pg部分蛋白发生结合、结合程度及结合位点,为将来研究光敏剂与细胞、细菌结合位点的筛选提供参考。
中图分类号:
- R781.4
| [1] |
Aoyama N, Suzuki JI, Kobayashi N, et al. Associations among tooth loss, systemic inflammation and antibody titers to periodontal pathogens in Japanese patients with cardiovascular disease[J]. J Periodontal Res, 2018, 53(1):117-122.
doi: 10.1111/jre.12494 pmid: 29139559 |
| [2] | Papapanou PN, Sanz M, Budunneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions[J]. J Periodontol, 2018, 89(Suppl 1):173-182. |
| [3] |
Yan X, Lu H, Zhang L, et al. A three-year study on periodontal microorganisms of short locking-taper implants and adjacent teeth in patients with history of periodontitis[J]. J Dent, 2020, 95:103299.
doi: 10.1016/j.jdent.2020.103299 |
| [4] | 梁雨晴, 董秤均, 伍文彬. 牙龈卟啉单胞菌与阿尔茨海默病的相关性研究进展[J]. 中华神经医学杂志, 2020, 19(5):525-527. |
| [5] | 王春萌, 洪丽华, 张志民, 等. 牙龈卟啉单胞菌在消化系统恶性肿瘤中的作用及机制[J]. 华西口腔医学杂志, 2019, 37(5):521-526. |
| [6] |
Kou Y, Inaba H, Kato T, et al. Inflammatory responses of gingival epithelial cells stimulated with Porphyromonas gingivalis vesicles are inhibited by hop-associated polyphenols[J]. J Periodontol, 2008, 79(1):174-180.
doi: 10.1902/jop.2008.070364 |
| [7] |
Ikal R, Hasegawa Y, Izumigawa M, et al. Mfa4, an accessory protein of Mfa1 fimbriae, modulates fimbrial biogenesis, cell auto-aggregation, and biofilm formation in Porphyromonas gingivalis[J]. PLoS One, 2015, 10(10):e0129454
doi: 10.1371/journal.pone.0129454 |
| [8] |
Kato T, Kawai S, Nakano K, et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype[J]. Cell Microbiol, 2007, 9(3):753-765.
doi: 10.1111/cmi.2007.9.issue-3 |
| [9] |
Cieplik F, Deng D, Crielaard W, et al. Antimicrobial photodynamic therapy: what we know and what we don’t[J]. Crit Rev Microbiol, 2018, 44(5):571-589.
doi: 10.1080/1040841X.2018.1467876 pmid: 29749263 |
| [10] |
Kwiatkowski S, Knap B, Prielaard D, et al. Photodynamic therapy mechanisms, photosensitizers and combinations[J]. Biomed Pharmacother, 2018, 106:1098-1107.
doi: S0753-3322(18)34161-1 pmid: 30119176 |
| [11] |
Malik Z, Ladan H, Nitzan Y. Photodynamic inactivation of Gram-negative bacteria: problems and possible solutions[J]. J Photochem Photobiol B, 1992, 14(3):262-266.
doi: 10.1016/1011-1344(92)85104-3 |
| [12] |
Nitzan Y, Gutterman M, Malik Z, et al. Inactivation of gram-negative bacteria by photosensitized porphyrins[J]. Photochem Photobiol, 1992, 55(1):89-96.
pmid: 1534909 |
| [13] |
Malik Z, Hanania J, Nitzan Y. Bactericidal effects of photoactivated porphyrins: an alternative approach to antimicrobial drugs[J]. J Photochem Photobiol B, 1990, 5(3/4):281-293.
doi: 10.1016/1011-1344(90)85044-W |
| [14] |
Wang KK, Finlay JC, Busch TM, et al. Explicit dosimetry for photodynamic therapy: macroscopic singlet oxygen modeling[J]. J Biophotonics, 2010, 3(5/6):304-318.
doi: 10.1002/jbio.v3:5/6 |
| [15] |
Kuimove MK, Yahioglu G, Ogilby PR. Singlet oxygen in a cell: spatially dependent lifetimes and quenching rate constants[J]. J Am Chem Soc, 2009, 131(1):332-340.
doi: 10.1021/ja807484b |
| [16] |
Redmond RW, Kochevar IE. Spatially resolved cellular responses to singlet oxygen[J]. Photochem Photobiol, 2006, 82(5):1178-1186.
pmid: 16740059 |
| [17] |
Pourhajibagher M, Bahador A. Gene expression profiling of fimA gene encoding fimbriae among clinical isolates of Porphyromonas gingivalis in response to photo-activated disinfection therapy[J]. Photodiagnosis Photodyn Ther, 2017, 20:1-5.
doi: S1572-1000(17)30343-5 pmid: 28797828 |
| [18] |
Pourhajibagher M, Bahador A. Evaluation of the crystal structure of a fimbrillin (FimA) from Porphyromonas gingivalis as a therapeutic target for photo-activated disinfection with toluidine blue O[J]. Photodiagnosis Photodyn Ther, 2017, 17:98-102.
doi: S1572-1000(16)30199-5 pmid: 27890593 |
| [19] |
Pourhajibagher M, Bahador A. In silico identification of a therapeutic target for photo-activated disinfection with indocyanine green: modeling and virtual screening analysis of Arg-gingipain from Porphyromonas gingivalis[J]. Photodiagnosis Photodyn Ther, 2017, 18:149-154.
doi: S1572-1000(17)30006-6 pmid: 28254551 |
| [20] |
Forli S, Huey R, Pique ME, et al. Computational protein-ligand docking and virtual drug screening with the AutoDock suite[J]. Nat Protoc, 2016, 11(5):905-919.
doi: 10.1038/nprot.2016.051 |
| [21] |
Ongarora BG, Fontenot KR, HU X, et al. Phthalocyanine-peptide conjugates for epidermal growth factor receptor targeting[J]. J Med Chem, 2012, 55(8):3725-3738.
doi: 10.1021/jm201544y pmid: 22468711 |
| [22] |
Shanmugaraj K, Anandakumar S, Ilanchelian M. Unraveling the binding interaction of Toluidine blue O with bovine hemoglobin: a multi spectroscopic and molecular modeling approach[J]. Rsc Advances, 2015, 5(6):3930-3940.
doi: 10.1039/C4RA11136B |
| [23] | Tsvetkov VB, Soloveva AB, Melik-Nubarov NS. Computer modeling of the complexes of Chlorin e6 with amphiphilic polymers[J]. Phys Chem Chem Phy, 2014, 16(22):10903-10913. |
| [24] |
Zhang Y, Wan Y, Chen Y, et al. Ultrasound-enhanced chemo-photodynamic combination therapy by using albumin “nanoglue”-based nanotheranostics[J]. Acs Nano, 2020, 14(5):5560-5569.
doi: 10.1021/acsnano.9b09827 |
| [25] |
Kandoussi I, Lakhlili W, Taoufik J, et al. Docking analysis of verteporfin with YAP WW domain[J]. Bioinformation, 2017, 13(7):237-240.
doi: 10.6026/97320630013237 pmid: 28943729 |
| [26] |
Alves E, Faustino MA, Neves MG, et al. An insight on bacterial cellular targets of photodynamic inactivation[J]. Future Med Chem, 2014, 6(2):141-164.
doi: 10.4155/fmc.13.211 |
| [27] |
Nagano K. FimA fimbriae of the periodontal disease-associated bacterium Porphyromonas gingivalis[J]. Yakugaku Zasshi, 2013, 133(9):963-974.
pmid: 23995804 |
| [28] |
Xu Q, Shoji M, Shibata S, et al. A distinct type of pilus from the human microbiome[J]. Cell, 2016, 165(3):690-703.
doi: 10.1016/j.cell.2016.03.016 |
| [29] |
Zhou XY, Gao JL, Hunter N, et al. Sequence-independent processing site of the C-terminal domain (CTD) influences maturation of the RgpB protease from Porphyromonas gingivalis[J]. Mol Microbiol, 2013, 89(5):903-917.
doi: 10.1111/mmi.2013.89.issue-5 |
| [30] | 尧晨光, 奚彩丽, 朱祥, 等. EV71 3C蛋白酶表达纯化、活性分析及与抑制剂模拟对接研究[J]. 中国病原生物学杂志, 2017, 12(8):722-726. |
| [31] | 刘福和, 陈少军, 倪文娟. 川芎中抗血栓活性成分的计算机虚拟筛选研究[J]. 中国药房, 2017, 28(16):2182-2186. |
| [1] | 曹泽,王乐童,刘振明. 严重急性呼吸综合征冠状病毒2的Spike蛋白点突变后与受体蛋白质及潜在抗病毒药物结合能力的同源建模分析[J]. 北京大学学报(医学版), 2021, 53(1): 150-158. |
| [2] | 轩艳,蔡宇,王啸轩,石巧,邱立新,栾庆先. 牙龈卟啉单胞菌感染对载脂蛋白e基因敲除小鼠动脉粥样硬化的影响[J]. 北京大学学报(医学版), 2020, 52(4): 743-749. |
| [3] | 高丽,于晓潜,蔡宇. 丝线结扎及局部涂抹牙龈卟啉单胞菌对小鼠牙槽骨骨吸收的影响[J]. 北京大学学报(医学版), 2017, 49(1): 31-035. |
| [4] | 李蓬1,万蒙2,刘健如2,李良忠1,张大鹍3△. 过氧化物酶体增生物激活受体γ在牙龈卟啉单胞菌刺激血管内皮细胞氧化应激中的作用[J]. 北京大学学报(医学版), 2015, 47(6): 977-982. |
| [5] | 王衣祥,安娜,欧阳翔英. 尼古丁和牙龈卟啉单胞菌脂多糖影响单核细胞黏附血管内皮细胞的分子机制[J]. 北京大学学报(医学版), 2015, 47(5): 809-813. |
| [6] | 吴迪, 欧阳翔英, 万蒙. 牙龈卟啉单胞菌感染内皮细胞导致白介素-8表达升高的胞内机制初探[J]. 北京大学学报(医学版), 2014, 46(2): 278-283. |
| [7] | 路瑞芳, 冯琳, 高学军, 孟焕新, 冯向辉. 侵袭性牙周炎龈沟液中有机酸与牙龈卟啉单胞菌和齿垢密螺旋体的关系[J]. 北京大学学报(医学版), 2013, 45(1): 12-16. |
| [8] | 谢静, 曾强, 郑扬, 廖锋, 徐国恒, 唐朝枢, 耿彬. 活细胞硫化氢检测新方法[J]. 北京大学学报(医学版), 2013, 45(03): 489-492. |
|
||