北京大学学报(医学版) ›› 2019, Vol. 51 ›› Issue (5): 805-812. doi: 10.19723/j.issn.1671-167X.2019.05.003
环状RNA circ-SOD2对肠上皮屏障和溃疡性结肠炎的作用
- 1. 北京大学人民医院 中心实验室, 北京 100044
2. 北京大学人民医院 消化内科,北京 100044
Effects of circular RNA circ-SOD2 on intestinal epithelial barrier and ulcerative colitis
Ting-ting WANG1,2,Ying HAN1,Fang-fang GAO1,Lei YE1,Yu-jun ZHANG1,△( )
)
- 1. The Central Laboratory, Peking University People’s Hospital, Beijing, 100044, China
2. Department of Gastroenterology, Peking University People’s Hospital, Beijing, 100044, China
摘要:
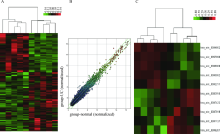
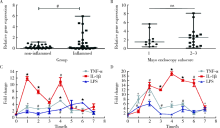
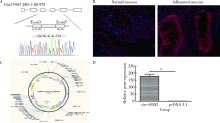
目的:探索溃疡性结肠炎(ulcerative colitis, UC)中环状RNA的表达谱改变,寻找在溃疡性结肠炎中表达发生明显异常的环状RNA,并探讨其对肠上皮细胞屏障功能的影响。方法:挑选5对UC患者炎症结直肠黏膜组织和正常黏膜组织进行环状RNA,芯片检测,筛选溃疡性结肠炎中表达发生改变的环状RNA。使用实时荧光定量PCR在30例UC患者炎症结直肠黏膜组织和正常黏膜组织中进一步验证表达发生明显上调的环状RNA circ-SOD2。使用炎症因子(LPS、TNF-α和IL1-β)刺激肠上皮细胞系Caco2、NMC460,检测circ-SOD2的表达改变。使用荧光原位杂交(fluorescence in situ hybridization, FISH)实验检测circ-SOD2在UC肠黏膜组织中的细胞定位。构建并合成circ-SOD2过表达载体,将其转染至Caco2细胞后检测Caco2细胞的跨上皮细胞电阻、FITC-右旋糖酐透过率,使用Western blotting检测上皮细胞屏障相关蛋白的表达改变。结果:环状RNA芯片检测后,通过限定条件(差异倍数>1.5,P<0.05), 在溃疡性结肠炎受累的结直肠黏膜中共筛选出111个上调、153个下调的circRNA。限定筛选条件:(1)在circRNA表达谱芯片中原始信号值(raw data)>100;(2)差异表达倍数>2倍;(3)差异有统计学意义(P<0.05),筛选出10个差异明显的circRNA,并最终锁定在UC炎症肠黏膜组织中上调倍数最高的circ-SOD2。进一步扩大样本量在30对溃疡性结肠炎患者炎症和正常肠黏膜组织中使用荧光实时定量PCR技术进行验证,发现circ-SOD2表达明显上调(P<0.001);使用LPS、TNF-α、IL1-β刺激Caco2、NCM460细胞后,发现circ-SOD2在刺激后1~7 h的不同时间点均表达上调。荧光原位杂交实验表明circ-SOD2主要表达于肠黏膜组织的肠上皮细胞,而在间质和炎症细胞中表达较少。在Caco2细胞中过表达circ-SOD2后,跨上皮细胞电阻明显下降,FITC-右旋糖酐透过率明显上升,上皮细胞屏障分子闭合蛋白(claudin-8, CLDN-8)表达明显下降(P<0.05)。结论:溃疡性结肠炎中环状RNA表达出现明显异常,其中在UC中表达上调的circ-SOD2可减弱肠上皮屏障,进而可能导致溃疡性结肠炎发生。
中图分类号:
- R574.62
| [1] | Magro FGP, Eliakim R, Ardizzone S , et al. Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders[J]. J Crohns Colitis, 2017,11(6):649-670. |
| [2] | Ng SC, Shi HY, Hamidi N , et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies[J]. Lancet, 2017,390(10114):2769-2778. |
| [3] | Costello CM, Mah N, Hasler R , et al. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays[J]. PLoS Med, 2005,2(8):e199. |
| [4] | Ventham NT, Kennedy NA, Nimmo ER , et al. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics[J]. Gastroenterology, 2013,145(2):293-308. |
| [5] | Jeck WR, Sorrentino JA, Wang K , et al. Circular RNAs are abundant, conserved, and associated with ALU repeats[J]. RNA, 2013,19(2):141-157. |
| [6] | Salzman J, Chen RE, Olsen MN , et al. Cell-type specifc features of circular RNA expression[J]. PLoS Genet, 2013,9(9):e1003777. |
| [7] | Han D, Li J, Wang H , et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression[J]. Hepatology, 2017,66(4):1151-1164. |
| [8] | Guarnerio J, Bezzi M, Jeong JC , et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations[J]. Cell, 2016,165(2):289-302. |
| [9] | Liu Q, Zhang X, Hu X , et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation[J]. Sci Rep, 2016,6:22572. |
| [10] | Iparraguirre L, Munoz-Culla M, Prada-Luengo I , et al. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis[J]. Hum Mol Genet, 2017,26(18):3564-3572. |
| [11] | Qiao YQ, Cai CW, Shen J , et al. Circular RNA expression alterations in colon tissues of Crohn’s disease patients[J]. Mol Med Rep, 2019,19(5):4500-4506. |
| [12] | Yuan G, Chen T, Zhang H , et al. Comprehensive analysis of differential circular RNA expression in a mouse model of colitis-induced colon carcinoma[J]. Mol Carcinog, 2018,57(12):1825-1834. |
| [13] | Piwecka M, Glazar P, Hernandez-Miranda LR , et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function[J]. Science, 2017,357(6357):8526. |
| [14] | Min M, Peng L, Yang Y , et al. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a[J]. Inflamm Bowel Dis, 2014,20(4):652-659. |
| [15] | Wang H, Chao K, Ng SC , et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease[J]. Genome Biol, 2016,17:58. |
| [16] | He C, Yu T, Shi Y , et al. MicroRNA 301A promotes intestinal inflammation and colitis-associated cancer development by inhi-biting BTG1[J]. Gastroenterology, 2017,152(6):1434-1448. |
| [17] | Qu S, Yang X, Li X , et al. Circular RNA: a new star of non-coding RNAs[J]. Cancer Lett, 2015,365(2):141-148. |
| [18] | Glažar P, Papavasileiou P, Rajewsky N . circBase: a database for circular RNAs[J]. RNA, 2014,20(11):1666-1670. |
| [19] | Kramer MC, Liang D, Tatomer DC , et al. Combinatorial control of drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins[J]. Genes Dev, 2015,29(20):2168-2182. |
| [20] | Gerasimenko TN, Senyavina NV, Anisimov NU , et al. A model of cadmium uptake and transport in Caco-2 cells[J]. Bull Exp Biol Med, 2016,161(1):187-192. |
| [21] | Mirza AH, Berthelsen CH, Seemann SE , et al. Transcriptomic landscape of lncRNAs in inflammatory bowel disease[J]. Genome Med, 2015,7(1):39. |
| [22] | Wu F, Huang Y, Dong F , et al. Ulcerative colitis-associated long noncoding RNA, BC012900, regulates intestinal epithelial cell apoptosis[J]. Inflamm Bowel Dis, 2016,22(4):782-795. |
| [23] | Hammond SM . An overview of microRNAs[J]. Adv Drug Deliv Rev, 2015,87:3-14. |
| [1] | 史佳琪,马莺,张奕,陈章健,贾光. 纳米二氧化钛颗粒对人肝癌细胞HepG2中circRNA表达谱的影响[J]. 北京大学学报(医学版), 2023, 55(3): 392-399. |
| [2] | 许云屹,苏征征,郑林茂,张孟尼,谭珺娅,杨亚蓝,张梦鑫,徐苗,陈铌,陈雪芹,周桥. 转录通读环状RNA rt-circ-HS促进低氧诱导因子1α表达和肾癌细胞增殖与侵袭[J]. 北京大学学报(医学版), 2023, 55(2): 217-227. |
| [3] | 贺冰洁,刘志科,沈鹏,孙烨祥,陈彬,詹思延,林鸿波. 2011—2020年宁波市鄞州区炎症性肠病发病的流行病学研究[J]. 北京大学学报(医学版), 2022, 54(3): 511-519. |
|
||