北京大学学报(医学版) ›› 2022, Vol. 54 ›› Issue (3): 532-540. doi: 10.19723/j.issn.1671-167X.2022.03.020
脓毒症小鼠髓源性抑制细胞氨基酸代谢特点
马媛1,张玥2,3,李蕊2,3,邓书伟2,3,秦秋实1,朱鏐娈1,2,3,*( )
)
- 1. 北京大学地坛医院教学医院传染病研究所,北京 100015
2. 首都医科大学附属北京地坛医院传染病研究所,北京 100015
3. 新发突发传染病研究北京市重点实验室,北京 100015
Characteristics of amino acid metabolism in myeloid-derived suppressor cells in septic mice
Yuan MA1,Yue ZHANG2,3,Rui LI2,3,Shu-wei DENG2,3,Qiu-shi QIN1,Liu-luan ZHU1,2,3,*( )
)
- 1. Institute of Infectious Diseases, Peking University Ditan Teaching Hospital, Beijing 100015, China
2. Institute of Infectious Diseases, Beijing Ditan Hospital, Capital Medical University, Beijing 100015, China
3. Beijing Key Laboratory of Emerging Infectious Diseases, Beijing 100015, China
摘要:
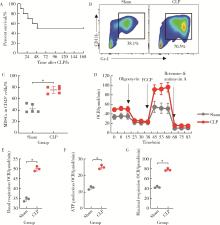
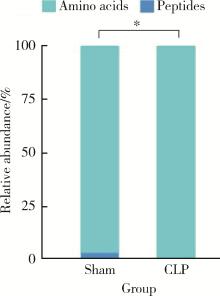
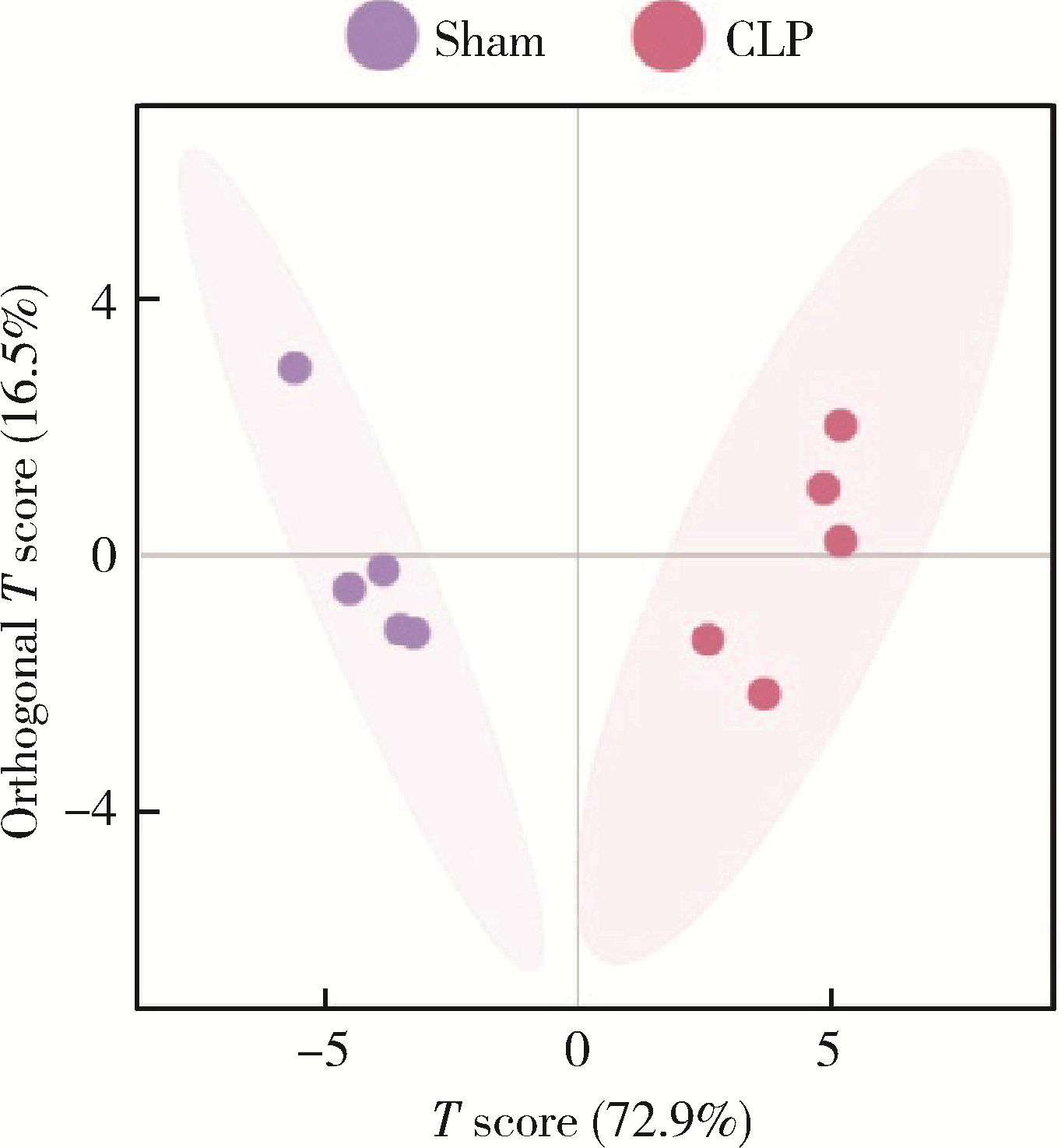
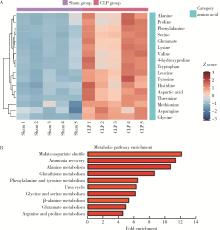
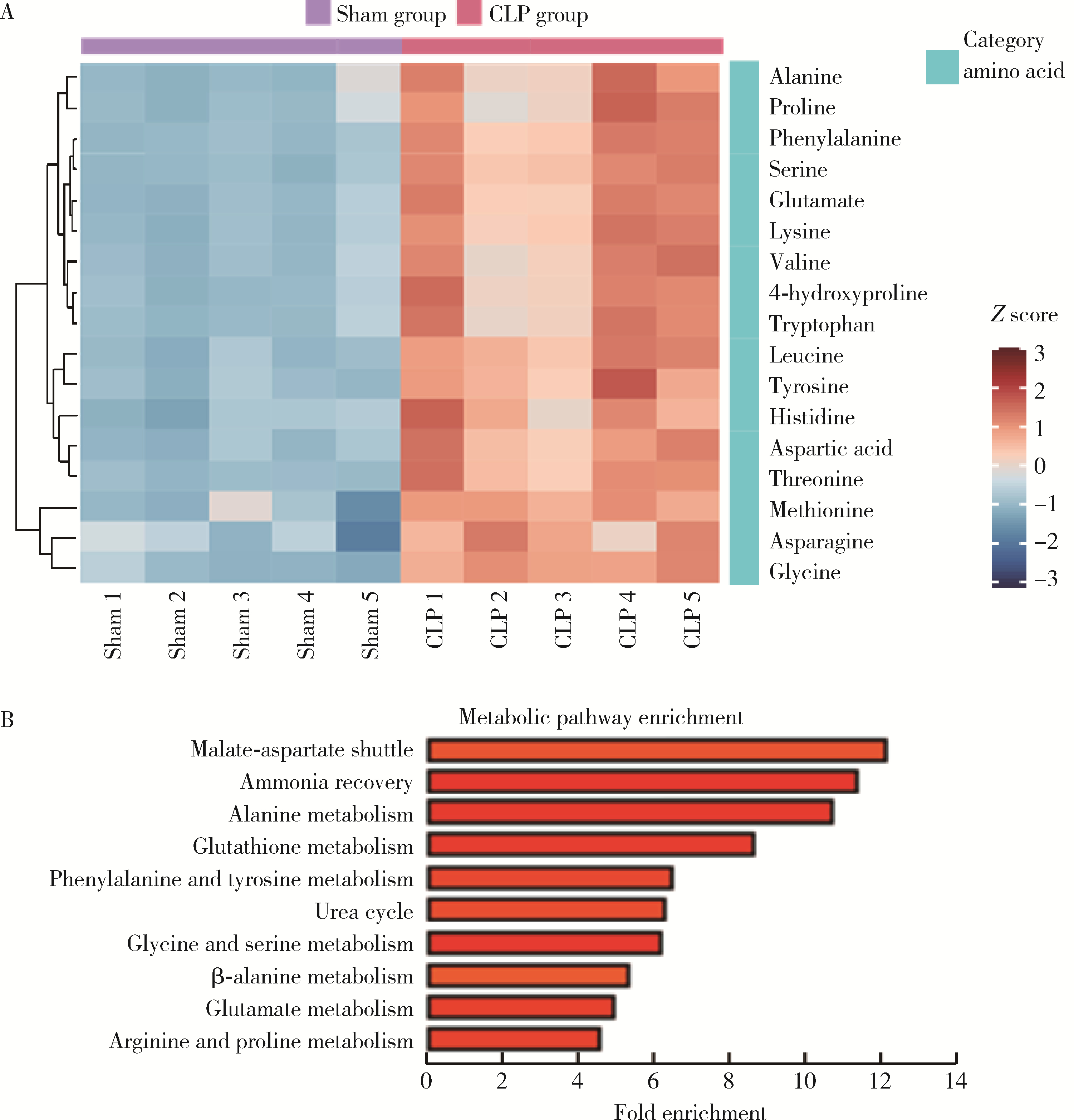
目的: 探讨脓毒症小鼠髓源性抑制细胞(myeloid-derived suppressor cells, MDSCs)氨基酸代谢组学特点。方法: 采用盲肠结扎穿孔术(cecal ligation and puncture, CLP)制备脓毒症小鼠模型,将小鼠随机分为假手术组(sham组, n = 10)和CLP组(n = 10)。术后第7天在各组存活小鼠中随机选取5只,分离小鼠骨髓MDSCs,采用安捷伦Seahorse XF技术测量MDSCs细胞的氧气消耗速率(oxygen consumption rate, OCR),采用超高效液相色谱-串联质谱联用技术靶向检测细胞内氨基酸及寡肽含量。通过单维和多维检验分析差异代谢物和潜在生物标志物,并对潜在生物标志物进行通路富集分析。结果: CLP组小鼠骨髓中MDSCs比例(75.53% ± 6.02%)显著大于sham组的MDSCs比例(43.15% ± 7.42%, t = 7.582, P < 0.001),且CLP组小鼠骨髓中MDSCs的基础呼吸速率[(50.03±1.20) pmol/min]、最大呼吸速率[(78.07±2.57) pmol/min]和腺嘌呤核苷三磷酸(adenosine triphosphate, ATP)产生[(25.30±1.21) pmol/min]均显著大于sham组的基础呼吸速率[(34.53±0.96) pmol/min, (t = 17.41, P < 0.001)]、最大呼吸速率[(42.57±1.87) pmol/min, (t = 19.33, P < 0.001)]和ATP产生[(12.63±0.96) pmol/min, (t = 14.18, P < 0.001)]。亮氨酸、苏氨酸、甘氨酸等17种氨基酸含量显著增加(均P < 0.05),是脓毒症MDSCs的潜在生物标志物。增加的氨基酸主要富集在苹果酸-天冬氨酸穿梭、氨回收、丙氨酸代谢、谷胱甘肽代谢、苯丙氨酸和酪氨酸代谢、尿素循环、甘氨酸和丝氨酸代谢、β-丙氨酸代谢、谷氨酸代谢、精氨酸和脯氨酸代谢等代谢途径。结论: CLP组小鼠MDSCs中线粒体氧化磷酸化增强,苹果酸-天冬氨酸穿梭和丙氨酸代谢增强,可能为线粒体有氧呼吸提供大量原料,从而促进MDSCs发挥免疫抑制功能,阻断上述代谢途径或将有助于调节MDSCs功能,为改善脓毒症预后提供新思路。
中图分类号:
- R363.1
| 1 |
Singer M , Deutschman CS , Seymour CW , et al. The third international consensus definitions for sepsis and septic shock (sepsis-3)[J]. JAMA, 2016, 315 (8): 801- 810.
doi: 10.1001/jama.2016.0287 |
| 2 |
徐静媛, 邱海波. 脓毒症治疗的现状与展望[J]. 国际流行病学传染病学杂志, 2021, 48 (4): 259- 262.
doi: 10.3760/cma.j.cn331340-20210618-00126 |
| 3 |
Rudd KE , Johnson SC , Agesa KM , et al. Global, regional, and national sepsis incidence and mortality, 1990—2017: analysis for the Global Burden of Disease Study[J]. Lancet, 2020, 395 (10219): 200- 211.
doi: 10.1016/S0140-6736(19)32989-7 |
| 4 |
Schrijver IT , Théroude C , Roger T . Myeloid-derived suppressor cells in sepsis[J]. Front Immunol, 2019, 10, 327.
doi: 10.3389/fimmu.2019.00327 |
| 5 |
Mira JC , Gentile LF , Mathias BJ , et al. Sepsis pathophysiology, chronic critical illness, and persistent inflammation-immuno-suppression and catabolism syndrome[J]. Crit Care Med, 2017, 45 (2): 253- 262.
doi: 10.1097/CCM.0000000000002074 |
| 6 |
Veglia F , Perego M , Gabrilovich D . Myeloid-derived suppressor cells coming of age[J]. Nat Immunol, 2018, 19 (2): 108- 119.
doi: 10.1038/s41590-017-0022-x |
| 7 |
Consonni FM , Porta C , Marino A , et al. Myeloid-Derived suppressor cells: ductile targets in disease[J]. Front Immunol, 2019, 10, 949.
doi: 10.3389/fimmu.2019.00949 |
| 8 |
Karin N . The Development and homing of myeloid-derived suppressor cells: from a two-stage model to a multistep narrative[J]. Front Immunol, 2020, 11, 557- 586.
doi: 10.3389/fimmu.2020.00557 |
| 9 |
Venet F , Monneret G . Advances in the understanding and treatment of sepsis-induced immunosuppression[J]. Nat Rev Nephrol, 2018, 14 (2): 121- 137.
doi: 10.1038/nrneph.2017.165 |
| 10 |
Mathias B , Delmas AL , Ozrazgat-Baslanti T , et al. Human myeloid-derived suppressor cells are associated with chronic immune suppression after severe sepsis/septic shock[J]. Ann Surg, 2017, 265 (4): 827- 834.
doi: 10.1097/SLA.0000000000001783 |
| 11 |
Darden DB , Bacher R , Brusko MA , et al. Single-cell RNA-seq of human myeloid-derived suppressor cells in late sepsis reveals multiple subsets with unique transcriptional responses: a pilot study[J]. Shock, 2021, 55 (5): 587- 595.
doi: 10.1097/SHK.0000000000001671 |
| 12 |
Wegiel B , Vuerich M , Daneshmandi S , et al. Metabolic switch in the tumor microenvironment determines immune responses to anti-cancer therapy[J]. Front Oncol, 2018, 8, 284.
doi: 10.3389/fonc.2018.00284 |
| 13 |
Won WJ , Deshane JS , Leavenworth JW , et al. Metabolic and functional reprogramming of myeloid-derived suppressor cells and their therapeutic control in glioblastoma[J]. Cell Stress, 2019, 3 (2): 47- 65.
doi: 10.15698/cst2019.02.176 |
| 14 |
Leinwand J , Miller G . Regulation and modulation of antitumor immunity in pancreatic cancer[J]. Nat Immunol, 2020, 21 (10): 1152- 1159.
doi: 10.1038/s41590-020-0761-y |
| 15 |
Grzywa TM , Sosnowska A , Matryba P , et al. Myeloid cell-derived arginase in cancer immune response[J]. Front Immunol, 2020, 11, 938.
doi: 10.3389/fimmu.2020.00938 |
| 16 |
Rittirsch D , Huber-Lang MS , Flierl MA , et al. Immunodesign of experimental sepsis by cecal ligation and puncture[J]. Nat Protoc, 2009, 4 (1): 31- 36.
doi: 10.1038/nprot.2008.214 |
| 17 |
Su L , Li H , Xie A , et al. Dynamic changes in amino acid concentration profiles in patients with sepsis[J]. PLoS One, 2015, 10 (4): e0121933.
doi: 10.1371/journal.pone.0121933 |
| 18 |
Gunst J , Vanhorebeek I , Thiessen SE , et al. Amino acid supplements in critically ill patients[J]. Pharmacol Res, 2018, 130, 127- 131.
doi: 10.1016/j.phrs.2017.12.007 |
| 19 |
Al-Khami AA , Rodriguez PC , Ochoa AC . Metabolic reprogramming of myeloid-derived suppressor cells (MDSCs) in cancer[J]. Oncoimmunology, 2016, 5 (8): e1200771.
doi: 10.1080/2162402X.2016.1200771 |
| 20 |
Mohammadpour H , MacDonald CR , McCarthy PL , et al. β2-adrenergic receptor signaling regulates metabolic pathways critical to myeloid-derived suppressor cell function within the TME[J]. Cell Rep, 2021, 37 (4): 109883.
doi: 10.1016/j.celrep.2021.109883 |
| 21 |
Altinok O , Poggio JL , Stein DE , et al. Malate-aspartate shuttle promotes l-lactate oxidation in mitochondria[J]. J Cell Physiol, 2020, 235 (3): 2569- 2581.
doi: 10.1002/jcp.29160 |
| 22 |
Mansouri S , Shahriari A , Kalantar H , et al. Role of malate dehydrogenase in facilitating lactate dehydrogenase to support the glycolysis pathway in tumors[J]. Biomed Rep, 2017, 6 (4): 463- 467.
doi: 10.3892/br.2017.873 |
| 23 |
Li X , Li Y , Yu Q , et al. Metabolic reprogramming of myeloid-derived suppressor cells: an innovative approach confronting challenges[J]. J Leukoc Biol, 2021, 110 (2): 257- 270.
doi: 10.1002/JLB.1MR0421-597RR |
| 24 |
Bozkus CC , Elzey BD , Crist SA , et al. Expression of cationic amino acid transporter 2 is required for myeloid-derived suppressor cell-mediated control of t cell immunity[J]. J Immunol, 2015, 195 (11): 5237- 5250.
doi: 10.4049/jimmunol.1500959 |
| 25 |
Srivastava MK , Sinha P , Clements VK , et al. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine[J]. Cancer Res, 2010, 70 (1): 68- 77.
doi: 10.1158/0008-5472.CAN-09-2587 |
| 26 |
Zhao H , Raines LN , Huang SC . Carbohydrate and amino acid metabolism as hallmarks for innate immune cell activation and function[J]. Cells, 2020, 9 (3): 562.
doi: 10.3390/cells9030562 |
| [1] | 曾媛媛,谢云,陈道南,王瑞兰. 脓毒症患者发生正常甲状腺性病态综合征的相关因素[J]. 北京大学学报(医学版), 2024, 56(3): 526-532. |
| [2] | 刘志伟,刘鹏,孟凡星,李天水,王颖,高嘉琪,周佐邑,王聪,赵斌. 内源性二氧化硫对脓毒症大鼠心肌氧化应激的调节[J]. 北京大学学报(医学版), 2023, 55(4): 582-586. |
| [3] | 刘余庆,卢剑,郝一昌,肖春雷,马潞林. 经皮肾镜取石术后尿脓毒血症的相关危险因素及预测模型[J]. 北京大学学报(医学版), 2018, 50(3): 507-513. |
| [4] | 张欣, 茹喜芳, 王颖, 李星, 桑田, 冯琪. 新生儿重症监护病房中新生儿真菌败血症的临床特点[J]. 北京大学学报(医学版), 2017, 49(5): 789-793. |
| [5] | 刘慧琳, 田勍, 洪天配, 刘桂花, 潘欢, 王海宁, 高洪伟. 脓毒症患者中血清程序化细胞死亡因子5水平的变化[J]. 北京大学学报(医学版), 2013, 45(2): 238-. |
|
||