Journal of Peking University (Health Sciences) ›› 2023, Vol. 55 ›› Issue (2): 217-227. doi: 10.19723/j.issn.1671-167X.2023.02.004
Previous Articles Next Articles
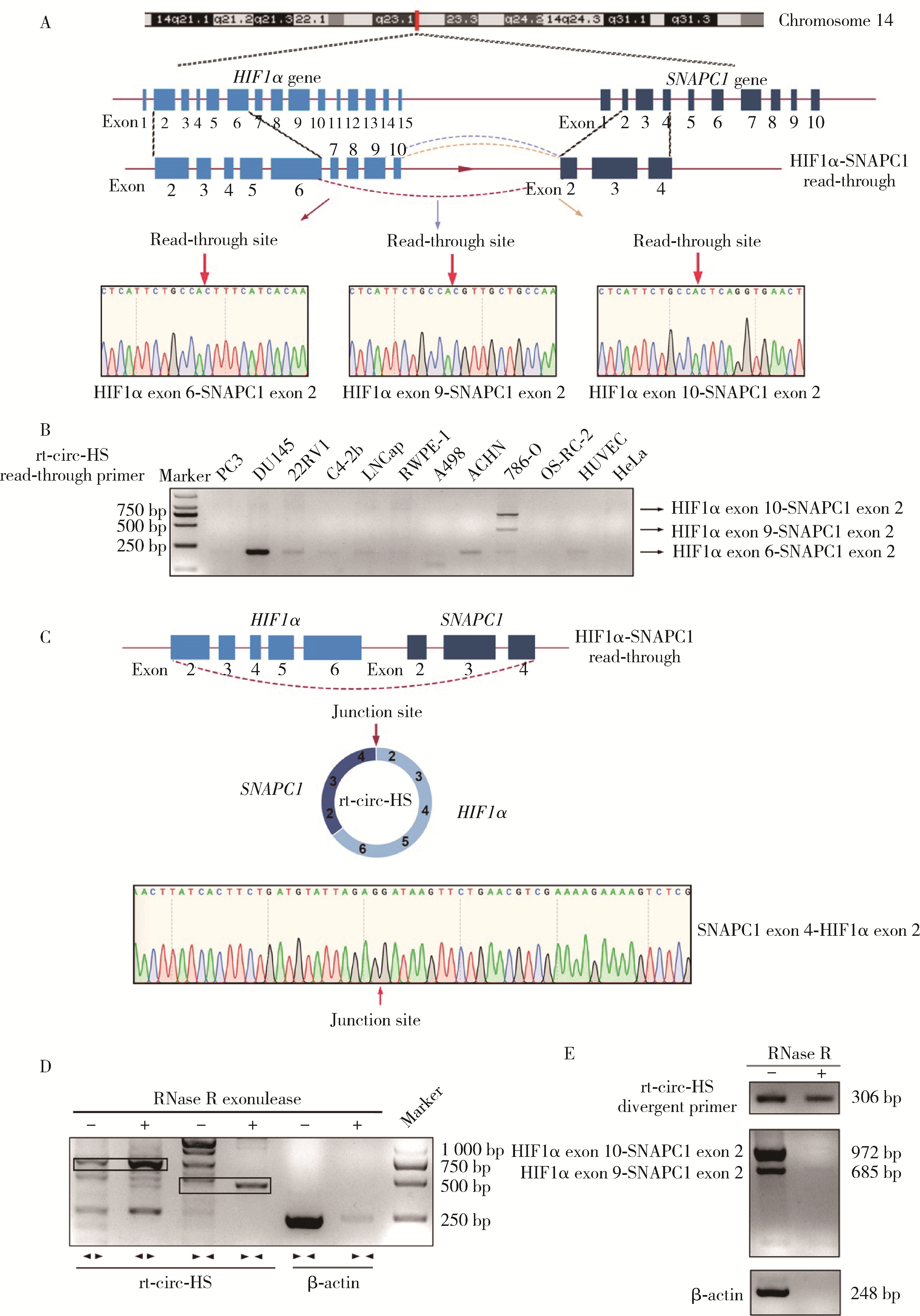
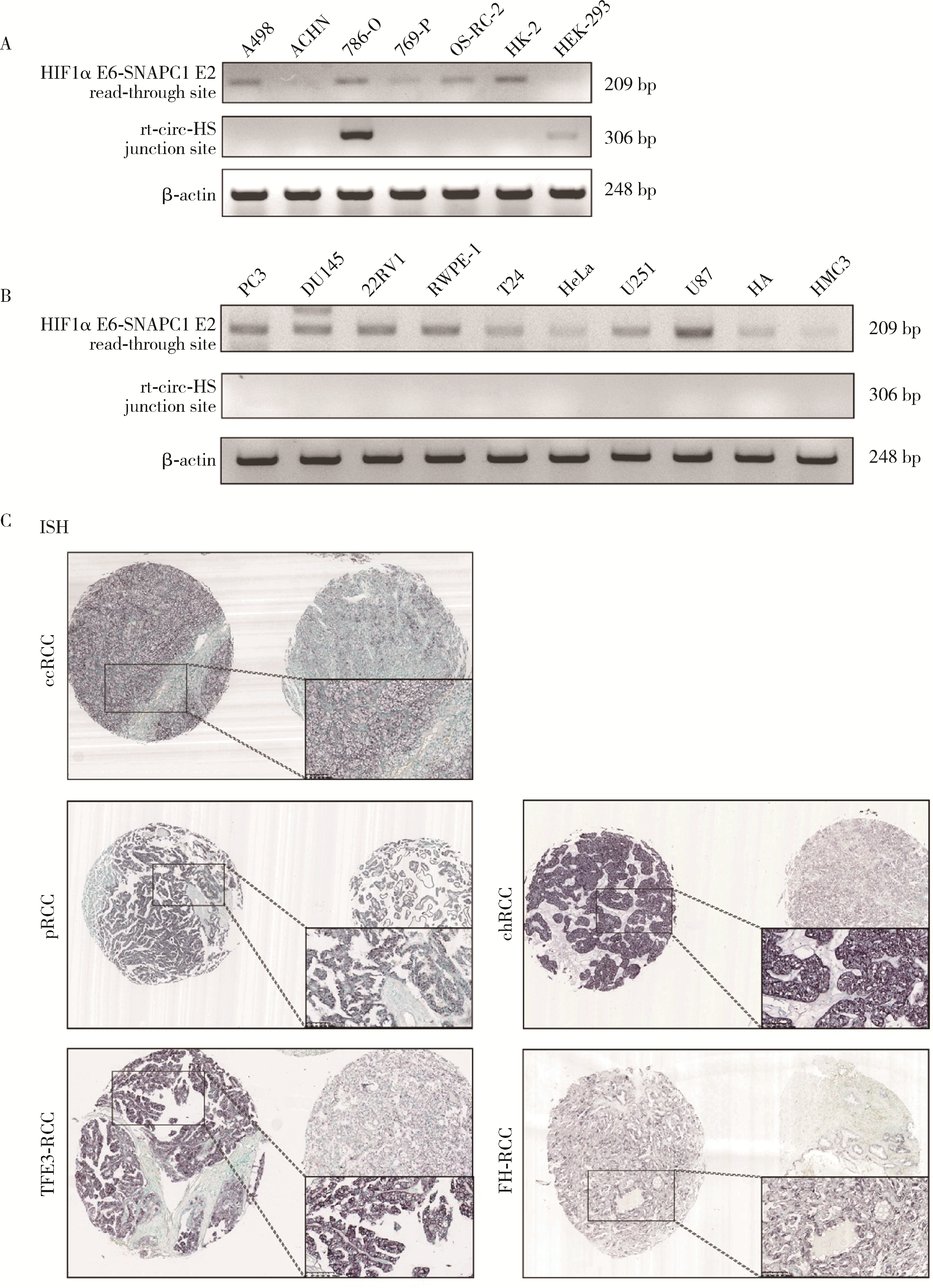
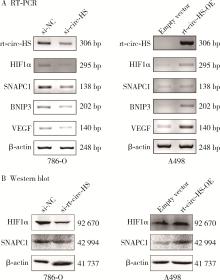
Read-through circular RNA rt-circ-HS promotes hypoxia inducible factor 1α expression and renal carcinoma cell proliferation, migration and invasiveness
Yun-yi XU1,Zheng-zheng SU1,Lin-mao ZHENG1,Meng-ni ZHANG1,Jun-ya TAN1,2,Ya-lan YANG1,Meng-xin ZHANG1,Miao XU1,Ni CHEN1,2,Xue-qin CHEN1,2,Qiao ZHOU1,2,*( )
)
- 1. Department of Pathology, West China Hospital, Sichuan University, Chengdu 610041, China
2. Research Laboratory of Pathology, West China Hospital, Sichuan University, Chengdu 610041, China
CLC Number:
- R365
| 1 |
Zhou WY , Cai ZR , Liu J , et al. Circular RNA: Metabolism, functions and interactions with proteins[J]. Mol Cancer, 2020, 19 (1): 172.
doi: 10.1186/s12943-020-01286-3 |
| 2 |
Chen Q , Liu T , Bao Y , et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway[J]. Cancer Lett, 2020, 469, 68- 77.
doi: 10.1016/j.canlet.2019.10.017 |
| 3 |
Mao W , Wang K , Xu B , et al. ciRS-7 is a prognostic biomarker and potential gene therapy target for renal cell carcinoma[J]. Mol Cancer, 2021, 20 (1): 142- 155.
doi: 10.1186/s12943-021-01443-2 |
| 4 |
van Zonneveld AJ , Kolling M , Bijkerk R , et al. Circular RNAs in kidney disease and cancer[J]. Nat Rev Nephrol, 2021, 17 (12): 814- 826.
doi: 10.1038/s41581-021-00465-9 |
| 5 |
Zhang Y , Gong M , Yuan H , et al. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation[J]. Cancer Discov, 2012, 2 (7): 598- 607.
doi: 10.1158/2159-8290.CD-12-0042 |
| 6 |
Grosso AR , Leite AP , Carvalho S , et al. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma[J]. Elife, 2015, 4, e09214.
doi: 10.7554/eLife.09214 |
| 7 |
Pflueger D , Mittmann C , Dehler S , et al. Functional characterization of BC039389-GATM and KLK4-KRSP1 chimeric read-through transcripts which are up-regulated in renal cell cancer[J]. BMC Genomics, 2015, 16 (1): 247.
doi: 10.1186/s12864-015-1446-z |
| 8 |
Chen N , Zhou Q . Constructing tissue microarrays without prefabricating recipient blocks: A novel approach[J]. Am J Clin Pathol, 2005, 124 (1): 103- 107.
doi: 10.1309/LHCJRFBUH8Q6QD3N |
| 9 |
Turajlic S , Swanton C , Boshoff C . Kidney cancer: The next decade[J]. J Exp Med, 2018, 215 (10): 2477- 2479.
doi: 10.1084/jem.20181617 |
| 10 |
Siegel RL , Miller KD , Fuchs HE , et al. Cancer statistics 2022[J]. CA Cancer J Clin, 2022, 72 (1): 7- 33.
doi: 10.3322/caac.21708 |
| 11 |
Garje R , Elhag D , Yasin HA , et al. Comprehensive review of chromophobe renal cell carcinoma[J]. Crit Rev Oncol Hematol, 2021, 160, 103287.
doi: 10.1016/j.critrevonc.2021.103287 |
| 12 |
Ji SQ , Su XL , Cheng WL , et al. Down-regulation of CD74 inhi-bits growth and invasion in clear cell renal cell carcinoma through HIF-1α pathway[J]. Urol Oncol, 2014, 32 (2): 153- 161.
doi: 10.1016/j.urolonc.2012.09.013 |
| 13 |
Hu CJ , Wang LY , Chodosh LA , et al. Differential roles of hypo-xia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation[J]. Mol Cell Biol, 2003, 23 (24): 9361- 9374.
doi: 10.1128/MCB.23.24.9361-9374.2003 |
| 14 |
Shen C , Beroukhim R , Schumacher SE , et al. Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene[J]. Cancer Discov, 2011, 1 (3): 222- 235.
doi: 10.1158/2159-8290.CD-11-0098 |
| 15 | Shinojima T , Oya M , Takayanagi A , et al. Renal cancer cells lacking hypoxia inducible factor (HIF)-1alpha expression maintain vascular endothelial growth factor expression through HIF-2alpha[J]. Carcinogenesis, 2007, 28 (3): 529- 536. |
| 16 |
Swiatek M , Jancewicz I , Kluebsoongnoen J , et al. Various forms of HIF-1alpha protein characterize the clear cell renal cell carcinoma cell lines[J]. IUBMB Life, 2020, 72 (6): 1220- 1232.
doi: 10.1002/iub.2281 |
| 17 |
Vidal AF . Read-through circular RNAs reveal the plasticity of RNA processing mechanisms in human cells[J]. RNA Biol, 2020, 17 (12): 1823- 1826.
doi: 10.1080/15476286.2020.1805233 |
| 18 |
Yang X , Ye T , Liu H , et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer[J]. Mol Cancer, 2021, 20 (1): 4.
doi: 10.1186/s12943-020-01300-8 |
| 19 |
Wu X , Zhou J , Zhao L , et al. CircCYP24A1 hampered malignant phenotype of renal cancer carcinoma through modulating CMTM-4 expression via sponging miR-421[J]. Cell Death Dis, 2022, 13 (2): 190.
doi: 10.1038/s41419-022-04623-0 |
| 20 |
Wang X , Xing L , Yang R , et al. The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC[J]. Mol Cancer, 2021, 20 (1): 91.
doi: 10.1186/s12943-021-01383-x |
| 21 |
Abdelmohsen K , Panda AC , Munk R , et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1[J]. RNA Biol, 2017, 14 (3): 361- 369.
doi: 10.1080/15476286.2017.1279788 |
| 22 |
Khan FA , Nsengimana B , Khan NH , et al. Chimeric peptides/proteins encoded by circRNA: An update on mechanisms and functions in human cancers[J]. Front Oncol, 2022, 12, 781270.
doi: 10.3389/fonc.2022.781270 |
| 23 |
Pintarelli G , Dassano A , Cotroneo CE , et al. Read-through transcripts in normal human lung parenchyma are down-regulated in lung adenocarcinoma[J]. Oncotarget, 2016, 7 (19): 27889- 27898.
doi: 10.18632/oncotarget.8556 |
| 24 |
Choi ES , Lee H , Lee CH , et al. Overexpression of KLHL23 protein from read-through transcription of PHOSPHO2-KLHL23 in gastric cancer increases cell proliferation[J]. FEBS Open Bio, 2016, 6 (11): 1155- 1164.
doi: 10.1002/2211-5463.12136 |
| 25 |
Wang L , Xiong X , Yao Z , et al. Chimeric RNA ASTN2-PAPPA(as) aggravates tumor progression and metastasis in human esophageal cancer[J]. Cancer Lett, 2021, 501, 1- 11.
doi: 10.1016/j.canlet.2020.10.052 |
| 26 |
Qin F , Zhang Y , Liu J , et al. SLC45A3-ELK4 functions as a long non-coding chimeric RNA[J]. Cancer Lett, 2017, 404, 53- 61.
doi: 10.1016/j.canlet.2017.07.007 |
| [1] | Fan SHU,Yichang HAO,Zhanyi ZHANG,Shaohui DENG,Hongxian ZHANG,Lei LIU,Guoliang WANG,Xiaojun TIAN,Lei ZHAO,Lulin MA,Shudong ZHANG. Functional and oncologic outcomes of partial nephrectomy for cystic renal cell carcinoma: A single-center retrospective study [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 667-672. |
| [2] | Zezhen ZHOU,Shaohui DENG,Ye YAN,Fan ZHANG,Yichang HAO,Liyuan GE,Hongxian ZHANG,Guoliang WANG,Shudong ZHANG. Predicting the 3-year tumor-specific survival in patients with T3a non-metastatic renal cell carcinoma [J]. Journal of Peking University (Health Sciences), 2024, 56(4): 673-679. |
| [3] | Yun-chong LIU,Zong-long WU,Li-yuan GE,Tan DU,Ya-qian WU,Yi-meng SONG,Cheng LIU,Lu-lin MA. Mechanism of nuclear protein 1 in the resistance to axitinib in clear cell renal cell carcinoma [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 781-792. |
| [4] | Dong LAN,Zhuo LIU,Yu-xuan LI,Guo-liang WANG,Xiao-jun TIAN,Lu-lin MA,Shu-dong ZHANG,Hong-xian ZHANG. Risk factors for massive hemorrhage after radical nephrectomy and removal of venous tumor thrombus [J]. Journal of Peking University (Health Sciences), 2023, 55(5): 825-832. |
| [5] | Qi SHEN,Yi-xiao LIU,Qun HE. Mucinous tubular and spindle cell carcinoma of kidney: Clinicopathology and prognosis [J]. Journal of Peking University (Health Sciences), 2023, 55(2): 276-282. |
| [6] | Quan ZHANG,Hai-feng SONG,Bing-lei MA,Zhe-nan ZHANG,Chao-hui ZHOU,Ao-lin LI,Jun LIU,Lei LIANG,Shi-yu ZHU,Qian ZHANG. Pre-operative prognostic nutritional index as a predictive factor for prognosis in non-metastatic renal cell carcinoma treated with surgery [J]. Journal of Peking University (Health Sciences), 2023, 55(1): 149-155. |
| [7] | Mei-ni ZUO,Yi-qing DU,Lu-ping YU,Xiang DAI,Tao XU. Correlation between metabolic syndrome and prognosis of patients with clear cell renal cell carcinoma [J]. Journal of Peking University (Health Sciences), 2022, 54(4): 636-643. |
| [8] | Tian-yu CAI,Zhen-peng ZHU,Chun-ru XU,Xing JI,Tong-de LV,Zhen-ke GUO,Jian LIN. Expression and significance of fibroblast growth factor receptor 2 in clear cell renal cell carcinoma [J]. Journal of Peking University (Health Sciences), 2022, 54(4): 628-635. |
| [9] | Er-shu BO,Peng HONG,Yu ZHANG,Shao-hui DENG,Li-yuan GE,Min LU,Nan LI,Lu-lin MA,Shu-dong ZHANG. Clinicopathological features and prognostic analysis of papillary renal cell carcinoma [J]. Journal of Peking University (Health Sciences), 2022, 54(4): 615-620. |
| [10] | Cai-peng QIN,Yu-xuan SONG,Meng-ting DING,Fei WANG,Jia-xing LIN,Wen-bo YANG,Yi-qing DU,Qing LI,Shi-jun LIU,Tao XU. Establishment of a mutation prediction model for evaluating the efficacy of immunotherapy in renal carcinoma [J]. Journal of Peking University (Health Sciences), 2022, 54(4): 663-668. |
| [11] | Yu TIAN,Xiao-yue CHENG,Hui-ying HE,Guo-liang WANG,Lu-lin MA. Clinical and pathological features of renal cell carcinoma with urinary tract tumor thrombus: 6 cases report and literature review [J]. Journal of Peking University (Health Sciences), 2021, 53(5): 928-932. |
| [12] | HONG Peng,TIAN Xiao-jun,ZHAO Xiao-yu,YANG Fei-long,LIU Zhuo,LU Min,ZHAO Lei,MA Lu-lin. Bilateral papillary renal cell carcinoma following kidney transplantation: A case report [J]. Journal of Peking University (Health Sciences), 2021, 53(4): 811-813. |
| [13] | HAN Song-chen,HUANG Zi-xiong,LIU Hui-xin,XU Tao. Renal functional compensation after unilateral radical nephrectomy of renal cell carcinoma [J]. Journal of Peking University (Health Sciences), 2021, 53(4): 680-685. |
| [14] | ZHAO Xun,YAN Ye,HUANG Xiao-juan,DONG Jing-han,LIU Zhuo,ZHANG Hong-xian,LIU Cheng,MA Lu-lin. Influence of deep invasive tumor thrombus on the surgical treatment and prognosis of patients with non-metastatic renal cell carcinoma complicated with venous tumor thrombus [J]. Journal of Peking University (Health Sciences), 2021, 53(4): 665-670. |
| [15] | SUN Zheng-hui,HUANG Xiao-juan,DONG Jing-han,LIU Zhuo,YAN Ye,LIU Cheng,MA Lu-lin. Risk factors of renal sinus invasion in clinical T1 renal cell carcinoma patients undergoing nephrectomy [J]. Journal of Peking University (Health Sciences), 2021, 53(4): 659-664. |
|
||