Journal of Peking University (Health Sciences) ›› 2025, Vol. 57 ›› Issue (5): 895-902. doi: 10.19723/j.issn.1671-167X.2025.05.013
Previous Articles Next Articles
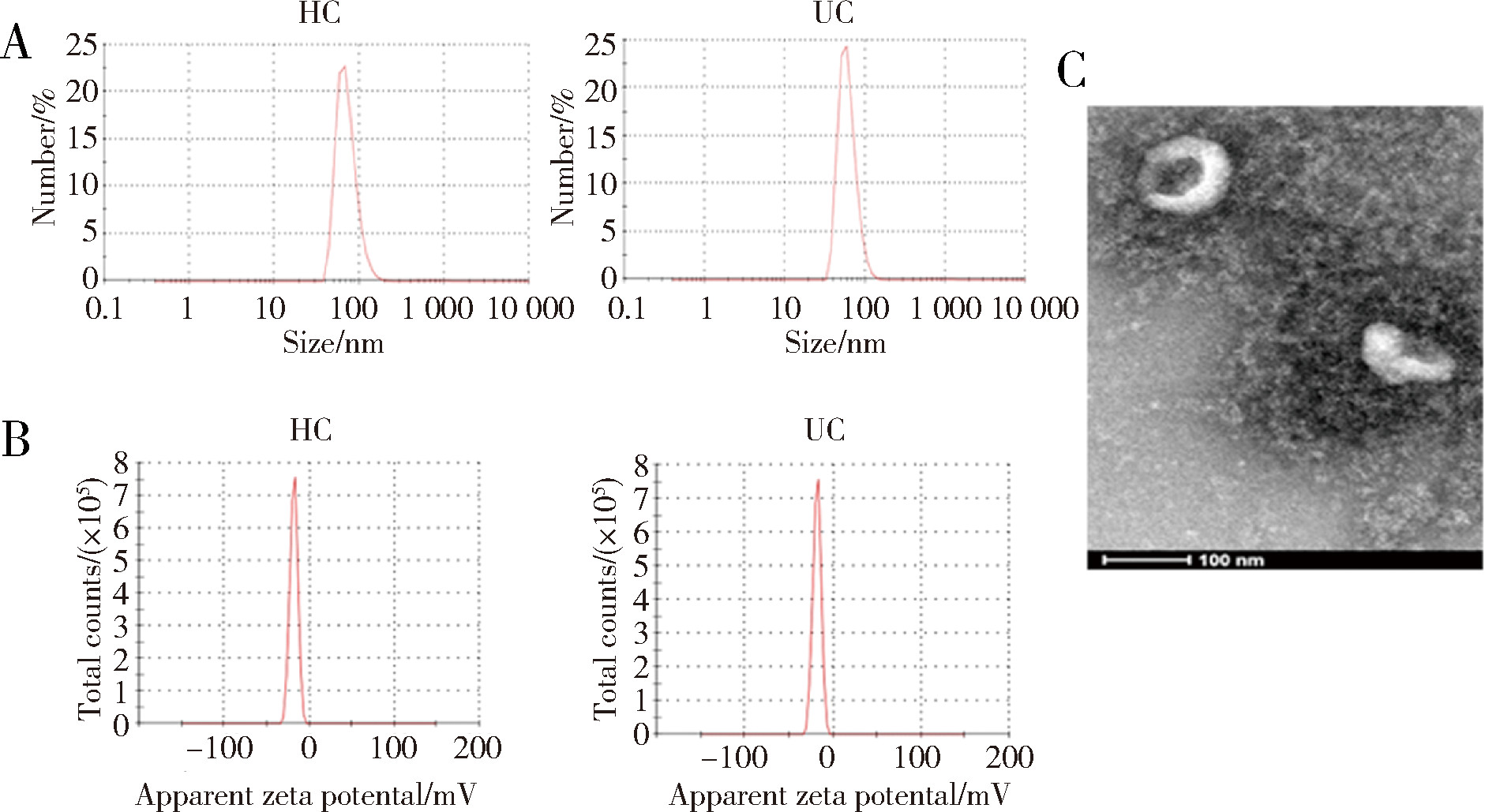
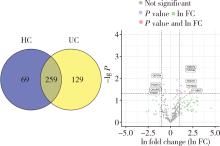
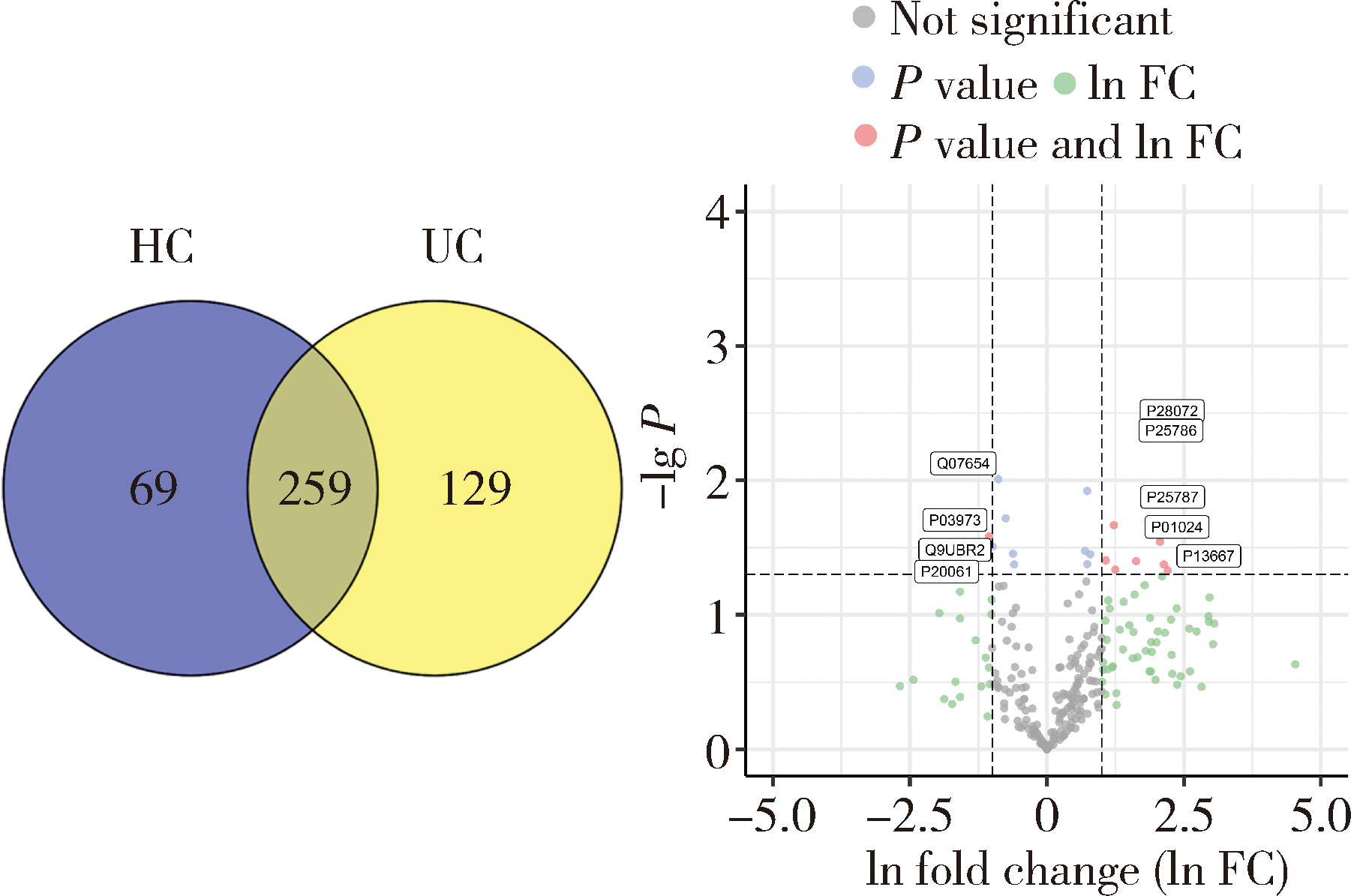
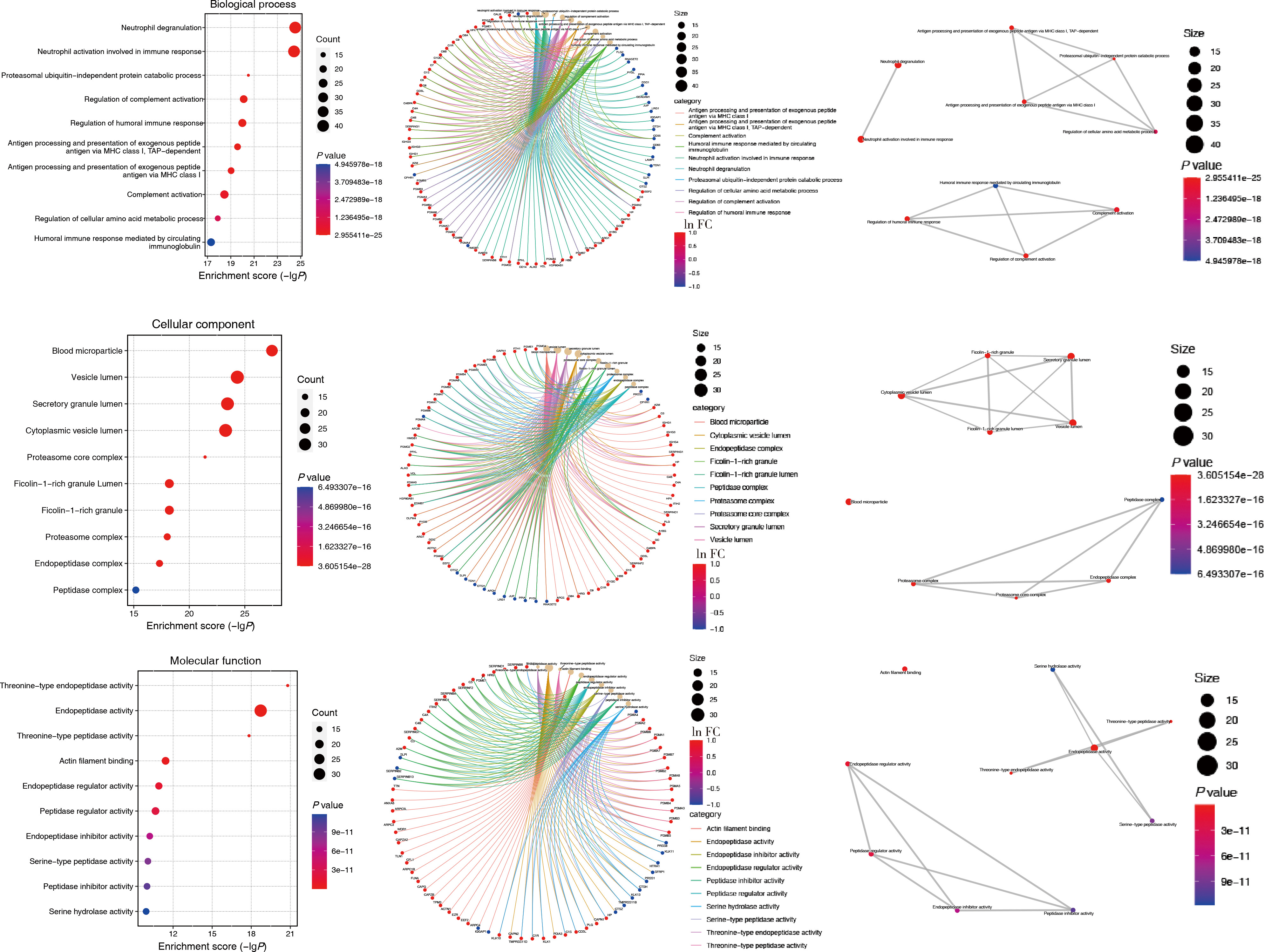
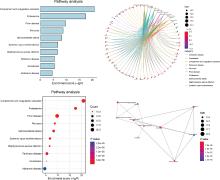
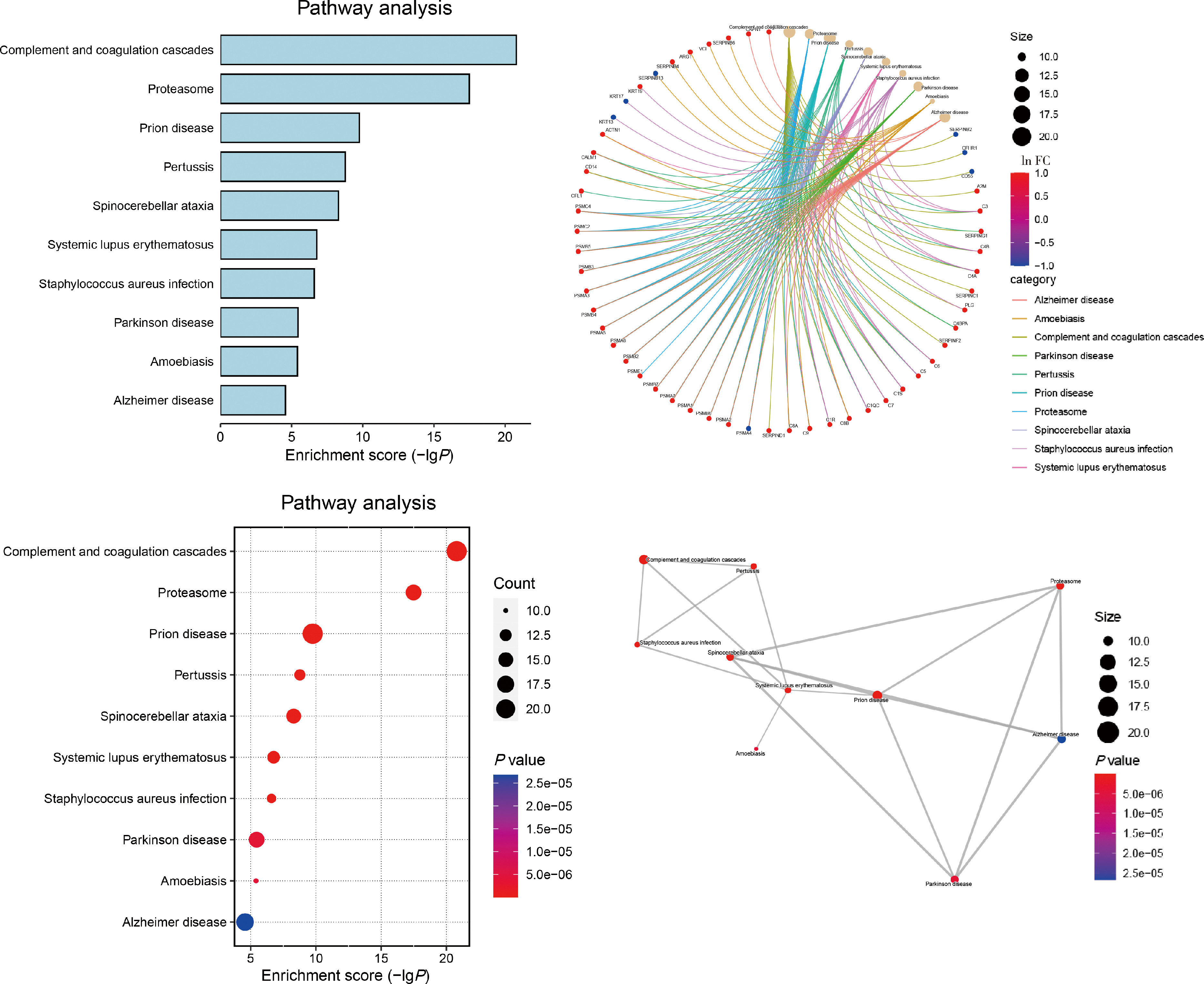
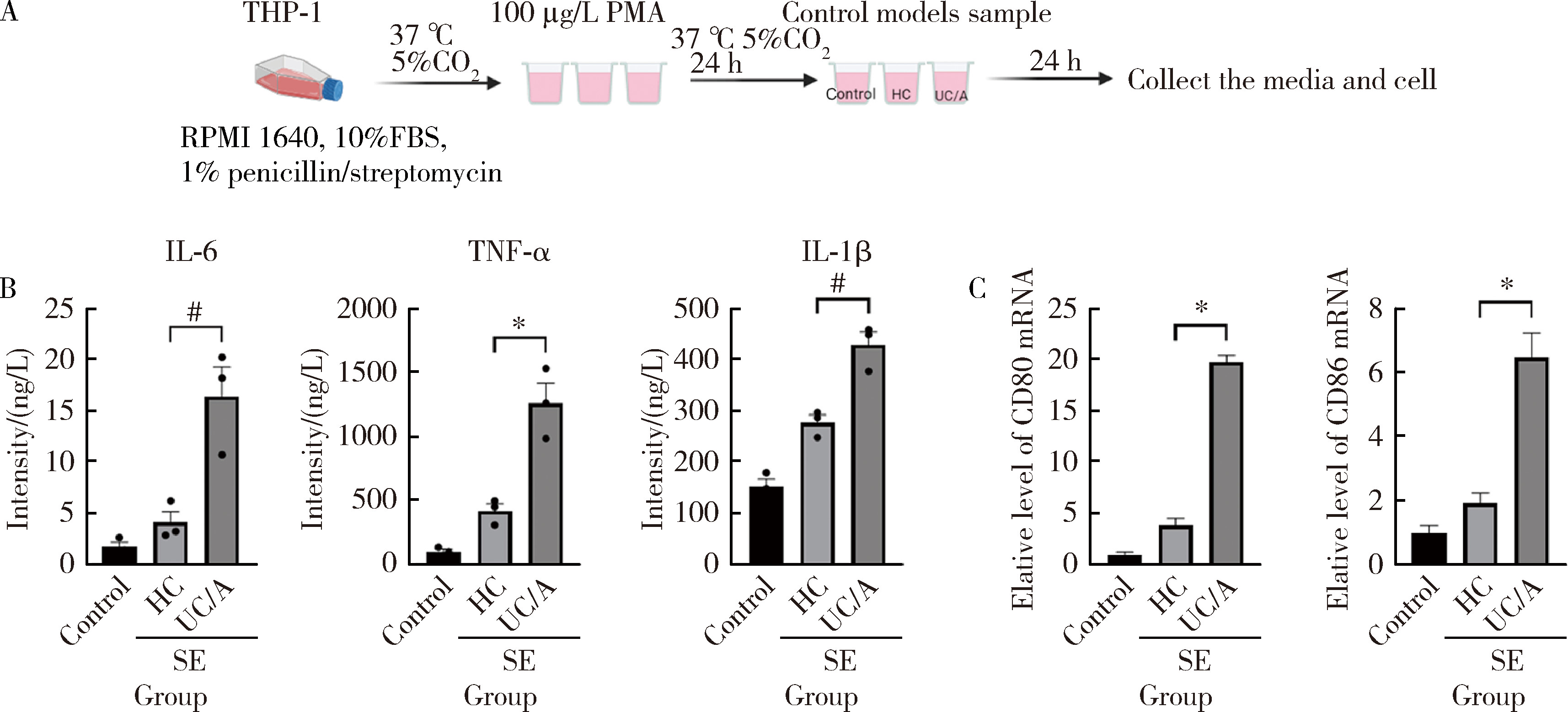
Protein biomarker screening and functional analysis of salivary exosomes in patients with ulcerative colitis
Congyi YANG1, Xiaowen ZHENG2, Jingyi CHEN1, Jun XU1, Feng CHEN2, Yang CHEN3, Ning CHEN1,*( )
)
- 1. Department of Gastroenterology, Peking University People's Hospital, Beijing 100044, China
2. Central Laboratory, Peking University School and Hospital of Stomatology & National Center of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Laboratory for Digital and Material Technology of Stomatology, Beijing 100081, China
3. Center for Precision Medicine Multi-Omics Research, Peking University School of Basic Medical Science, Beijing 100191, China
CLC Number:
- R574.62
| 1 |
|
| 2 |
doi: 10.1016/S0140-6736(17)32448-0 |
| 3 |
doi: 10.1038/nature06005 |
| 4 |
doi: 10.1136/gut.2005.069476 |
| 5 |
doi: 10.1126/science.aau6977 |
| 6 |
doi: 10.1073/pnas.1618088114 |
| 7 |
doi: 10.1002/adma.201802896 |
| 8 |
doi: 10.1089/omi.2011.0118 |
| 9 |
doi: 10.1093/bioinformatics/btt285 |
| 10 |
doi: 10.3390/cells8070727 |
| 11 |
doi: 10.1111/prd.12100 |
| 12 |
doi: 10.1016/j.arr.2022.101587 |
| 13 |
doi: 10.7150/ijbs.25018 |
| 14 |
|
| 15 |
doi: 10.1186/s11658-021-00280-x |
| 16 |
doi: 10.1016/j.stem.2012.11.009 |
| 17 |
doi: 10.1136/gutjnl-2012-303205 |
| 18 |
doi: 10.1186/s41232-016-0011-8 |
| 19 |
doi: 10.1038/nm.1978 |
| 20 |
doi: 10.1016/j.addr.2011.05.010 |
| 21 |
doi: 10.1002/pmic.201500174 |
| 22 |
doi: 10.1136/gut.30.3.361 |
| 23 |
doi: 10.1126/science.aan4526 |
| 24 |
doi: 10.1097/MIB.0000000000000840 |
| [1] | Xiangyu SUN, Chao YUAN, Xinzhu ZHOU, Jing DIAO, Shuguo ZHENG. Application of salivary micro-ecosystem in early prevention and control of oral and systemic diseases [J]. Journal of Peking University (Health Sciences), 2025, 57(5): 859-863. |
| [2] | Zhong CAO,Hong-bing CEN,Jian-hong ZHAO,Jun MEI,Ling-zhi QIN,Wei LIAO,Qi-lin AO. Expression and significance of INSM1 and SOX11 in pancreatic neuroendocrine tumor and solid pseudopapillary neoplasm [J]. Journal of Peking University (Health Sciences), 2023, 55(4): 575-581. |
| [3] | Fang CAO,Ming ZHONG,Cong-rong LIU. Uterine POLE mutant endometrioid carcinoma combined with human papilloma virus-associated cervical adenocarcinoma: A case report and literature review [J]. Journal of Peking University (Health Sciences), 2023, 55(2): 370-374. |
| [4] | Li LIANG,Xin LI,Lin NONG,Ying DONG,Ji-xin ZHANG,Dong LI,Ting LI. Analysis of microsatellite instability in endometrial cancer: The significance of minimal microsatellite shift [J]. Journal of Peking University (Health Sciences), 2023, 55(2): 254-261. |
| [5] | Tian-yu CAI,Zhen-peng ZHU,Chun-ru XU,Xing JI,Tong-de LV,Zhen-ke GUO,Jian LIN. Expression and significance of fibroblast growth factor receptor 2 in clear cell renal cell carcinoma [J]. Journal of Peking University (Health Sciences), 2022, 54(4): 628-635. |
| [6] | Bing-jie HE,Zhi-ke LIU,Peng SHEN,Ye-xiang SUN,Bin CHEN,Si-yan ZHAN,Hong-bo LIN. Epidemiological study on the incidence of inflammatory bowel disease in Yinzhou District, Ningbo City from 2011 to 2020 [J]. Journal of Peking University (Health Sciences), 2022, 54(3): 511-519. |
| [7] | PANG Yong,ZHANG Sha,YANG Hua,ZHOU Rou-li. Serum LAPTM4B-35 protein as a novel diagnostic marker for hepatocellular carcinoma [J]. Journal of Peking University (Health Sciences), 2021, 53(4): 710-715. |
| [8] | ZHOU Chuan, MA Xue, XING Yun-kun, LI Lu-di, CHEN Jie, YAO Bi-yun, FU Juan-ling, ZHAO Peng. Exploratory screening of potential pan-cancer biomarkers based on The Cancer Genome Atlas database [J]. Journal of Peking University (Health Sciences), 2021, 53(3): 602-607. |
| [9] | CHEN Huai-an,LIU Shuo,LI Xiu-jun,WANG Zhe,ZHANG Chao,LI Feng-qi,MIAO Wen-long. Clinical value of inflammatory biomarkers in predicting prognosis of patients with ureteral urothelial carcinoma [J]. Journal of Peking University (Health Sciences), 2021, 53(2): 302-307. |
| [10] | Xu-chu ZHANG,Jian-hua ZHANG,Rong-fu WANG,Yan FAN,Zhan-li FU,Ping YAN,Guang-yu ZHAO,Yan-xia BAI. Diagnostic value of 18F-FDG PET/CT and tumor markers (CEA, CA19-9, CA24-2) in recurrence and metastasis of postoperative colorectal moderately differentiated adenocarcinoma [J]. Journal of Peking University(Health Sciences), 2019, 51(6): 1071-1077. |
| [11] | Ting-ting WANG,Ying HAN,Fang-fang GAO,Lei YE,Yu-jun ZHANG. Effects of circular RNA circ-SOD2 on intestinal epithelial barrier and ulcerative colitis [J]. Journal of Peking University(Health Sciences), 2019, 51(5): 805-812. |
| [12] | Fei-long YANG,Kai HONG,Guo-jiang ZHAO,Cheng LIU,Yi-meng SONG,Lu-lin MA. Construction of prognostic model and identification of prognostic biomarkers based on the expression of long non-coding RNA in bladder cancer via bioinformatics [J]. Journal of Peking University(Health Sciences), 2019, 51(4): 615-622. |
| [13] | LIU Jin, XIONG Geng-yan, TANG Qi, FANG Dong, LI Xue-song, ZHOU Li-qun. Methylation status of RASSF1A gene promoter in upper tract urothelial carcinoma and its clinical significance [J]. Journal of Peking University(Health Sciences), 2016, 48(4): 571-578. |
|
||