北京大学学报(医学版) ›› 2020, Vol. 52 ›› Issue (5): 809-814. doi: 10.19723/j.issn.1671-167X.2020.05.003
中国人群Hedgehog通路基因与非综合征型唇腭裂的亲源效应
李文咏1,王梦莹1,周仁1,王斯悦1,郑鸿尘1,朱洪平2,周治波2,吴涛1,3,∆( ),王红1,石冰4
),王红1,石冰4
- 1.北京大学公共卫生学院流行病与卫生统计学系,北京 100191
2.北京大学口腔医学院·口腔医院,口腔颌面外科 国家口腔疾病临床医学研究中心 口腔数字化医疗技术和材料国家工程实验室 口腔数字医学北京市重点实验室,北京 100081
3.卫生部生育健康重点实验室,北京 100191
4.四川大学华西口腔医学院口腔颌面外科,口腔疾病研究国家重点实验室,成都 610041
Exploring parent-of-origin effects for non-syndromic cleft lip with or without cleft palate on PTCH1, PTCH2, SHH, SMO genes in Chinese case-parent trios
Wen-yong LI1,Meng-ying WANG1,Ren ZHOU1,Si-yue WANG1,Hong-chen ZHENG1,Hong-ping ZHU2,Zhi-bo ZHOU2,Tao WU1,3,∆( ),Hong WANG1,Bing SHI4
),Hong WANG1,Bing SHI4
- 1. Department of Epidemiology and Biostatistics, School of Public Health, Peking University, Beijing 100191, China
2. Department of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Laboratory for Digital and Material Technology of Stomatology & Beijing Key Laboratory of Digital Stomatology, Beijing 100081, China
3. Key Laboratory of Reproductive Health, Ministry of Health, Beijing 100191, China
4. Department of Oral and Maxillofacial Surgery, State Key Laboratory of Oral Disease, West China College of Stomatology, Sichuan University, Chengdu 610041, China
摘要:
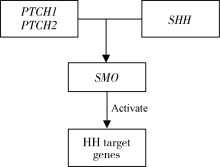
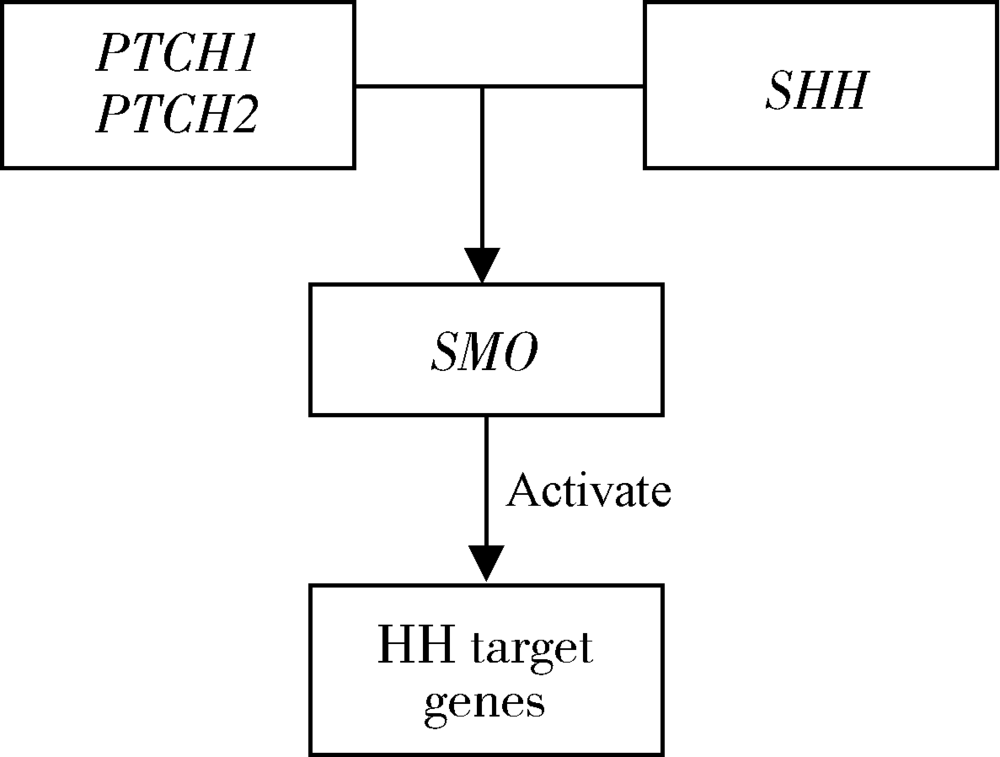
目的:探索非综合征型唇腭裂(non-syndromic cleft lip with or without cleft palate,NSCL/P)这一类常见出生缺陷的可能致病机制,在Hedgehog(HH)通路基因中(PTCH1、PTCH2、SHH、SMO)探索基因多态性对NSCL/P的关联关系以及亲源效应(parent-of-origin effects,PoO)对NSCL/P发病风险的影响。方法:纳入806个中国非综合征型唇腭裂核心家系,对HH通路基因(PTCH1、PTCH2、SHH、SMO)的83个单核苷酸多态性位点(single nucleotide polymorphisms,SNPs)进行传递不平衡检验(transmission disequilibrium test,TDT), 并采用对数线性模型进行亲源效应分析。家系样本来自“唇腭裂基因和交互作用的国际合作研究”项目。采用Plink进行TDT检验;通过R软件中的Haplin v6.2.1软件包开展亲源效应分析。采用Bonferroni法进行多重检验校正。结果:经过质量控制,共纳入65个SNPs进行分析,Bonferroni显著性水平为7.7×10 -4(0.05/65)。未校正P值前,关联分析发现rs4448343与NSCL/P存在关联(P=0.023), 6个单体型(rs10512249-rs4448343、rs1461208-rs7786445、rs10512249-rs4448343、rs16909865-rs10512249-rs4448343、rs1461208-rs7786445-rs12698335、rs288756-rs288758-rs1151790)与NSCL/P存在关联(P<0.05);6个单体型(rs288765-rs1233563、rs12537550-rs11765352、rs872723-rs288765-rs1233563、rs288765-rs1233563-rs288756、rs6459952-rs12537550-rs11765352、rs12537550-rs11765352-rs6971211)具有潜在的PoO效应(P<0.05)。以上结果经过多重检验校正,均无统计学意义(P>7.7×10 -4)。结论:未发现HH通路基因多态性与NSCL/P的关联,未发现HH通路基因通过PoO效应影响NSCL/P发病风险。
中图分类号:
- R394
| [1] |
Wang M, Yuan Y, Wang Z, et al. Prevalence of orofacial clefts among live births in China: a systematic review and meta-analysis[J]. Birth Defects Res, 2017,109(13):1011-1019.
doi: 10.1002/bdr2.1043 pmid: 28635078 |
| [2] |
Harville EW, Wilcox AJ, Lie RT, et al. Cleft lip and palate versus cleft lip only: are they distinct defects[J]. Am J Epidemiol, 2005,162(5):448-453.
doi: 10.1093/aje/kwi214 pmid: 16076837 |
| [3] |
Leslie EJ, Marazita ML. Genetics of cleft lip and cleft palate[J]. Am J Med Genet C Semin Med Genet, 2013,163c(4):246-258.
doi: 10.1002/ajmg.c.31381 pmid: 24124047 |
| [4] |
Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms[J]. Dev Dyn, 2006,235(5):1152-1166.
doi: 10.1002/dvdy.20646 pmid: 16292776 |
| [5] |
Mossey PA, Little J, Munger RG, et al. Cleft lip and palate[J]. Lancet, 2009,374(9703):1773-1785.
doi: 10.1016/S0140-6736(09)60695-4 pmid: 19747722 |
| [6] |
Grant SF, Wang K, Zhang H, et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24[J]. J Pediatr, 2009,155(6):909-913.
doi: 10.1016/j.jpeds.2009.06.020 pmid: 19656524 |
| [7] |
Beaty TH, Marazita ML, Leslie EJ. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow's opportunities[J]. F1000Res, 2016,5:2800.
doi: 10.12688/f1000research.9503.1 pmid: 27990279 |
| [8] |
Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases[J]. Nature, 2009,461(7265):747-753.
doi: 10.1038/nature08494 pmid: 19812666 |
| [9] |
Guilmatre A, Sharp AJ. Parent of origin effects[J]. Clin Genet, 2012,81(3):201-209.
doi: 10.1111/j.1399-0004.2011.01790.x |
| [10] |
Yu Y, Zuo X, He M, et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity[J]. Nat Commun, 2017,8:14364.
doi: 10.1038/ncomms14364 pmid: 28232668 |
| [11] |
Beaty TH, Murray JC, Marazita ML, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4[J]. Nat Genet, 2010,42(6):525-529.
doi: 10.1038/ng.580 pmid: 20436469 |
| [12] |
Gjessing HK, Lie RT. Case-parent triads: estimating single- and double-dose effects of fetal and maternal disease gene haplotypes[J]. Ann Hum Genet, 2006,70(Pt 3):382-396.
doi: 10.1111/j.1529-8817.2005.00218.x pmid: 16674560 |
| [13] |
Gjerdevik M, Haaland OA, Romanowska J, et al. Parent-of-origin-environment interactions in case-parent triads with or without independent controls[J]. Ann Hum Genet, 2018,82(2):60-73.
doi: 10.1111/ahg.12224 pmid: 29094765 |
| [14] |
Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads[J]. Am J Hum Genet, 1999,65(1):229-235.
doi: 10.1086/302466 pmid: 10364536 |
| [15] |
Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease[J]. Nat Rev Mol Cell Biol, 2013,14(7):416-429.
doi: 10.1038/nrm3598 |
| [16] |
Taipale J, Cooper MK, Maiti T, et al. Patched acts catalytically to suppress the activity of Smoothened[J]. Nature, 2002,418(6900):892-897.
doi: 10.1038/nature00989 pmid: 12192414 |
| [17] |
Wantia N, Rettinger G. The current understanding of cleft lip malformations[J]. Facial Plast Surg, 2002,18(3):147-153.
doi: 10.1055/s-2002-33061 pmid: 12152133 |
| [18] |
Grosen D, Bille C, Petersen I, et al. Risk of oral clefts in twins[J]. Epidemiology, 2011,22(3):313-319.
doi: 10.1097/EDE.0b013e3182125f9c |
| [19] |
Mangold E, Ludwig KU, Birnbaum S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate[J]. Nat Genet, 2010,42(1):24-26.
doi: 10.1038/ng.506 pmid: 20023658 |
| [20] |
Sun Y, Huang Y, Yin A, et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate[J]. Nat Commun, 2015,6:6414.
pmid: 25775280 |
| [21] |
Lo Muzio L. Nevoid basal cell carcinoma syndrome (Gorlin syndrome)[J]. Orphanet J Rare Dis, 2008,3(1):32.
doi: 10.1186/1750-1172-3-32 |
| [22] |
Metzis V, Courtney AD, Kerr MC, et al. Patched1 is required in neural crest cells for the prevention of orofacial clefts[J]. Hum Mol Genet, 2013,22(24):5026-5035.
doi: 10.1093/hmg/ddt353 |
| [23] |
Xiao Y, Taub MA, Ruczinski I, et al. Evidence for SNP-SNP interaction identified through targeted sequencing of cleft case-parent trios[J]. Genet Epidemiol, 2017,41(3):244-250.
pmid: 28019042 |
| [24] |
de Araujo TK, Secolin R, Felix TM, et al. A multicentric association study between 39 genes and nonsyndromic cleft lip and palate in a Brazilian population[J]. J Craniomaxillofac Surg, 2016,44(1):16-20.
pmid: 26602496 |
| [25] |
Rubini M, Brusati R, Garattini G, et al. Cystathionine beta-synthase c.844ins68 gene variant and non-syndromic cleft lip and palate[J]. Am J Med Genet A, 2005,136a(4):368-372.
pmid: 16007597 |
| [26] |
Reutter H, Birnbaum S, Mende M, et al. TGFB3 displays parent-of-origin effects among central Europeans with nonsyndromic cleft lip and palate[J]. J Hum Genet, 2008,53(7):656-661.
doi: 10.1007/s10038-008-0296-9 |
| [27] |
Sull JW, Liang KY, Hetmanski JB, et al. Differential parental transmission of markers in RUNX2 among cleft case-parent trios from four populations[J]. Genet Epidemiol, 2008,32(6):505-512.
pmid: 18357615 |
| [28] |
Sull JW, Liang KY, Hetmanski JB, et al. Maternal transmission effects of the PAX genes among cleft case-parent trios from four populations[J]. Eur J Hum Genet, 2009,17(6):831-839.
pmid: 19142206 |
| [29] |
Sull JW, Liang KY, Hetmanski JB, et al. Evidence that TGFA influences risk to cleft lip with/without cleft palate through unconventional genetic mechanisms[J]. Hum Genet, 2009,126(3):385-394.
pmid: 19444471 |
| [30] |
Suazo J, Santos JL, Jara L, et al. Parent-of-origin effects for MSX1 in a Chilean population with nonsyndromic cleft lip/palate[J]. Am J Med Genet A, 2010,152a(8):2011-2016.
pmid: 20635363 |
| [31] |
Shi M, Murray JC, Marazita ML, et al. Genome wide study of maternal and parent-of-origin effects on the etiology of orofacial clefts[J]. Am J Med Genet A, 2012,158a(4):784-794.
doi: 10.1002/ajmg.a.35257 |
| [32] |
Garg P, Ludwig KU, Bohmer AC, et al. Genome-wide analysis of parent-of-origin effects in non-syndromic orofacial clefts[J]. Eur J Hum Genet, 2014,22(6):822-830.
doi: 10.1038/ejhg.2013.235 |
| [33] |
Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles[J]. Genet Epidemiol, 2002,23(3):221-233.
pmid: 12384975 |
| [1] | 侯天姣,周治波,王竹青,王梦莹,王斯悦,彭和香,郭煌达,李奕昕,章涵宇,秦雪英,武轶群,郑鸿尘,李静,吴涛,朱洪平. 转化生长因子β信号通路与非综合征型唇腭裂发病风险的基因-基因及基因-环境交互作用[J]. 北京大学学报(医学版), 2024, 56(3): 384-389. |
| [2] | 王梦莹,李文咏,周仁,王斯悦,刘冬静,郑鸿尘,周治波,朱洪平,吴涛,胡永华. WNT信号通路基因位点单体型与中国汉族人群非综合征型唇腭裂发病风险的关联[J]. 北京大学学报(医学版), 2022, 54(3): 394-399. |
| [3] | 王梦莹,李文咏,周仁,王斯悦,刘冬静,郑鸿尘,李静,李楠,周治波,朱洪平,吴涛,胡永华. WNT代谢通路相关基因与中国人群非综合征型唇腭裂发病风险的交互作用[J]. 北京大学学报(医学版), 2020, 52(5): 815-820. |
| [4] | 周仁,郑鸿尘,李文咏,王梦莹,王斯悦,李楠,李静,周治波,吴涛,朱洪平. 利用二代测序数据探索SPRY基因家族与中国人群非综合征型唇腭裂的关联[J]. 北京大学学报(医学版), 2019, 51(3): 564-570. |
| [5] | 张杰铌,宋凤岐,周绍楠,郑晖,彭丽颖,张倩,赵望泓,张韬文,李巍然,周治波,林久祥,陈峰. 中国唇腭裂患者Sonic hedgehog信号通路相关单核苷酸多态性的分析[J]. 北京大学学报(医学版), 2019, 51(3): 556-563. |
|
||