北京大学学报(医学版) ›› 2025, Vol. 57 ›› Issue (3): 456-464. doi: 10.19723/j.issn.1671-167X.2025.03.008
肥胖指标与DNA甲基化时钟关系的纵向双生子研究
刘顺恺1,2, 曹卫华1,2, 吕筠1,2, 余灿清1,2, 黄涛1,2, 孙点剑一1,2, 廖春晓1,2, 庞元捷1,2, 胡润华1,2, 高汝钦3, 俞敏4, 周金意5, 吴先萍6, 刘彧7, 高文静1,2,*( ), 李立明1,2
), 李立明1,2
- 1. 北京大学公共卫生学院流行病与卫生统计学系, 北京 100191
2. 重大疾病流行病学教育部重点实验室(北京大学), 北京 100191
3. 青岛市疾病预防控制中心, 山东青岛 266033
4. 浙江省疾病预防控制中心, 杭州 310051
5. 江苏省疾病预防控制中心, 南京 210008
6. 四川省疾病预防控制中心, 成都 610041
7. 黑龙江省疾病预防控制中心, 哈尔滨 150090
Association between DNA methylation clock and obesity-related indicators: A longitudinal twin study
Shunkai LIU1,2, Weihua CAO1,2, Jun LV1,2, Canqing YU1,2, Tao HUANG1,2, Dianjianyi SUN1,2, Chunxiao LIAO1,2, Yuanjie PANG1,2, Runhua HU1,2, Ruqin GAO3, Min YU4, Jinyi ZHOU5, Xianping WU6, Yu LIU7, Wenjing GAO1,2,*( ), Liming LI1,2
), Liming LI1,2
- 1. Department of Epidemiology and Biostatistics, Peking University School of Public Health, Beijing 100191, China
2. Key Laboratory of Epidemiology of Major Diseases (Peking University), Ministry of Education, Beijing 100191, China
3. Qingdao Center for Disease Control and Prevention, Qingdao 266033, Shandong, China
4. Zhejiang Center for Disease Control and Prevention, Hangzhou 310051, China
5. Jiangsu Center for Disease Control and Prevention, Nanjing 210008, China
6. Sichuan Center for Disease Control and Prevention, Chengdu 610041, China
7. Heilongjiang Center for Disease Control and Prevention, Harbin 150090, China
摘要:
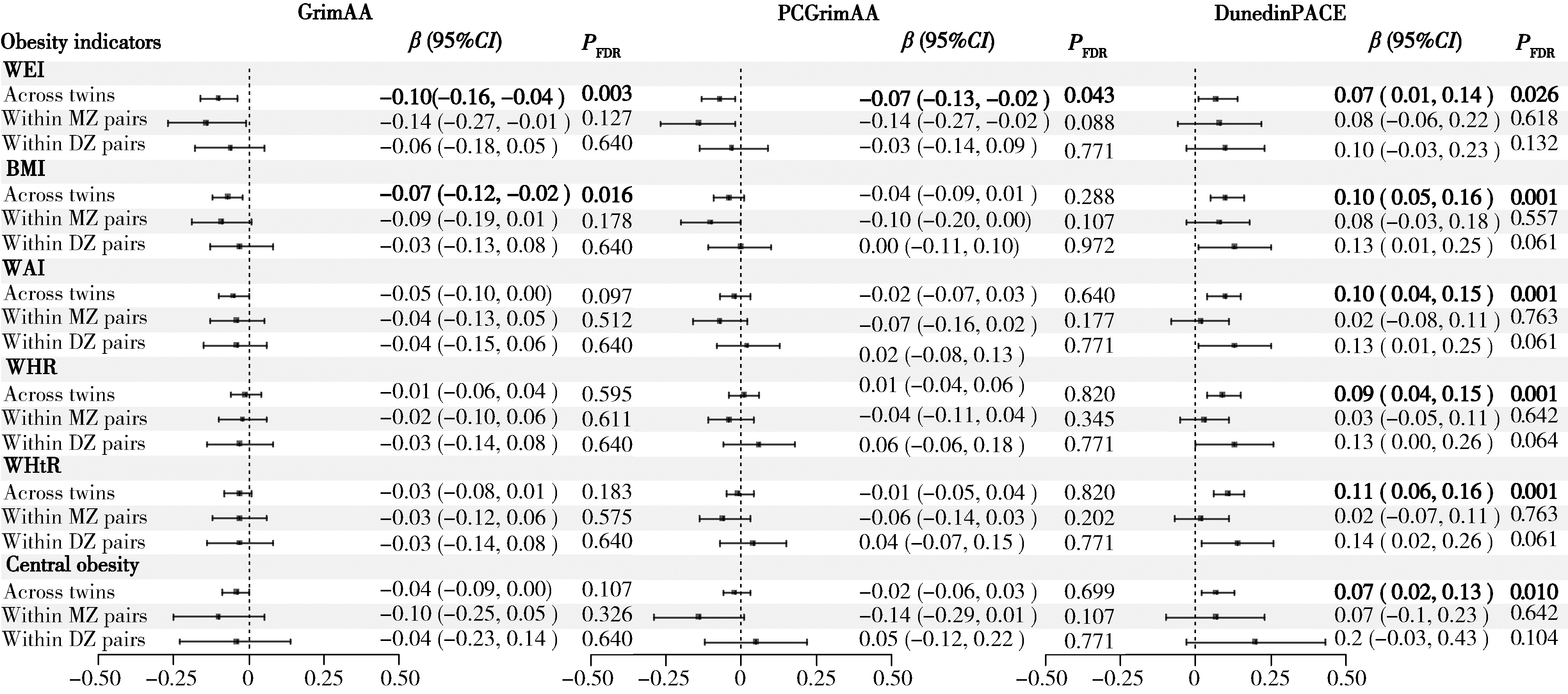
目的: 探讨肥胖相关指标与DNA甲基化时钟及其加速指标之间的关系,并分析两者在时间上的先后顺序。方法: 研究数据来源于中国双生子登记系统在2013年和2017—2018年开展的两次专题调查。通过Illumina Infinium人类甲基化450K芯片和EPIC芯片测定外周血DNA甲基化数据,并采用DNA甲基化年龄计算器(https://dnamage.genetics.ucla.edu/)或研究者提供的R代码计算DNA甲基化时钟指标GrimAA、PCGrimAA和DunedinPACE。肥胖指标包括体重、体重指数(body mass index,BMI)、腰围、腰臀比、腰高比。横断面分析纳入1 070名双生子,对内分析同卵双生子378对,异卵双生子155对,采用混合效应模型分析肥胖指标与DNA甲基化时钟及其加速指标的相关性。纵向分析纳入314名双生子,对内分析同卵双生子95对,异卵双生子62对,采用交叉滞后模型进一步探索肥胖与DNA甲基化时钟指标间的时间顺序关联。上述分析均分别在全部双生子和同卵、异卵双生子对内进行。结果: 横断面分析人群中同卵双生子占71.0%,男性占68.0%,平均实足年龄为(49.9±12.1)岁;纵向分析人群中同卵双生子占60.5%,男性占60.8%;基线平均实足年龄为(50.4±10.2)岁,平均随访时间(4.6±0.6)年。除随访时腰臀比均值高于基线外,其他肥胖指标基线与随访均值差异无统计学意义。相关性分析显示,在全部双生子中,体重、BMI、腰围、腰臀比、腰高比均与DunedinPACE时钟呈正相关,其中腰高比与DunedinPACE时钟的关联最为显著(β=0.21,95%CI:0.11~0.31);体重和BMI均与GrimAA呈负相关(β=-0.03,95%CI:-0.05~-0.01; β=-0.07,95%CI:-0.12~-0.02),体重与PCGrimAA呈负相关(β=-0.02,95%CI:-0.03~0.00);但双生子对内分析中的相关性未达到统计学显著水平。交叉滞后分析显示,基线体重升高可能引起随访时GrimAA增加,基线体重、BMI和腰围升高可能引起随访时PCGrimAA增加,基线腰臀比升高可能引起随访时DunedinPACE升高。结论: 肥胖指标与DNA甲基化时钟指标存在相关,基线肥胖指标对随访时部分DNA甲基化时钟指标的变化具有影响,肥胖可能通过加速DNA甲基化时钟和衰老进程对个体健康产生长期影响,但二者间的关联受双生子共享的遗传或环境因素影响。
中图分类号:
- R181.33
| 1 |
doi: 10.1038/s41576-022-00511-7 |
| 2 |
doi: 10.1093/gerona/gls233 |
| 3 |
doi: 10.18632/aging.101684 |
| 4 |
doi: 10.7554/eLife.73420 |
| 5 |
doi: 10.1038/s43587-022-00248-2 |
| 6 |
doi: 10.3389/fgene.2022.819749 |
| 7 |
doi: 10.1073/pnas.2215840120 |
| 8 |
doi: 10.1111/obr.13724 |
| 9 |
doi: 10.3390/ijms23169252 |
| 10 |
doi: 10.1038/s41574-021-00551-9 |
| 11 |
doi: 10.1016/j.freeradbiomed.2018.09.039 |
| 12 |
doi: 10.1093/ajcn/nqy107 |
| 13 |
|
| 14 |
doi: 10.1002/oby.23255 |
| 15 |
doi: 10.1093/aje/kwaa251 |
| 16 |
doi: 10.1111/joim.12926 |
| 17 |
中华人民共和国卫生和计划生育委员会. WS/T428-2013成人体重判定[S]. 北京: 中国标准出版社, 2013.
|
| 18 |
doi: 10.1371/journal.pone.0123992 |
| 19 |
doi: 10.1093/gerona/glz099 |
| 20 |
doi: 10.3390/ijms20174273 |
| 21 |
doi: 10.1161/CIRCRESAHA.119.315397 |
| 22 |
doi: 10.1038/s41598-022-11562-5 |
| 23 |
doi: 10.1186/s12944-024-02042-y |
| 24 |
doi: 10.1111/joim.13528 |
| 25 |
doi: 10.1038/s41366-024-01466-x |
| 26 |
doi: 10.1016/j.arr.2011.12.003 |
| 27 |
doi: 10.1111/obr.12991 |
| 28 |
doi: 10.1113/JP271691 |
| 29 |
doi: 10.1016/j.bcp.2021.114723 |
| [1] | 张依航, 蔡珊, 陈子玥, 刘云飞, 党佳佳, 师嫡, 李佳欣, 黄天彧, 宋逸. 基于RE-AIM框架儿童青少年近视与肥胖共病综合干预实施性研究结局指标的构建[J]. 北京大学学报(医学版), 2025, 57(3): 436-441. |
| [2] | 李萍, 王海雪, 高晓, 韩亚静, 王辉, 王海俊, 牟莹莹. 基于移动健康技术对超重或肥胖孕妇体重管理的随机对照试验[J]. 北京大学学报(医学版), 2025, 57(3): 465-472. |
| [3] | 刘慧丽, 闻蓓, 白雪, 陈明安, 李民. 体重校正腰围指数与疼痛的相关性:一项横断面研究[J]. 北京大学学报(医学版), 2025, 57(1): 178-184. |
| [4] | 陈敬,单蕊,肖伍才,张晓蕊,刘峥. 青春期和成年早期自制力与抑郁症状和超重肥胖共病风险的关联:基于全国调查的十年前瞻性队列研究[J]. 北京大学学报(医学版), 2024, 56(3): 397-402. |
| [5] | 吴一凡,玉应香,谢岚,张志达,常翠青. 不同体重指数青年男性的静息能量消耗特点及预测方程评价[J]. 北京大学学报(医学版), 2024, 56(2): 247-252. |
| [6] | 陈楚云,孙蓬飞,赵静,贾佳,范芳芳,王春燕,李建平,姜一梦,霍勇,张岩. 北京社区人群促红细胞生成素相关因素及其与10年心血管疾病风险的关系[J]. 北京大学学报(医学版), 2023, 55(6): 1068-1073. |
| [7] | 党佳佳,蔡珊,钟盼亮,王雅琪,刘云飞,师嫡,陈子玥,张依航,胡佩瑾,李晶,马军,宋逸. 室外夜间人工光暴露与中国9~18岁儿童青少年超重肥胖的关联[J]. 北京大学学报(医学版), 2023, 55(3): 421-428. |
| [8] | 陈敬,肖伍才,单蕊,宋洁云,刘峥. DRD2基因rs2587552多态性对儿童肥胖干预效果的影响:一项前瞻性、平行对照试验[J]. 北京大学学报(医学版), 2023, 55(3): 436-441. |
| [9] | 马涛,李艳辉,陈曼曼,马莹,高迪,陈力,马奇,张奕,刘婕妤,王鑫鑫,董彦会,马军. 青春期启动提前与儿童肥胖类型的关联研究: 基于横断面调查和队列调查[J]. 北京大学学报(医学版), 2022, 54(5): 961-970. |
| [10] | 娄雪,廖莉,李兴珺,王楠,刘爽,崔若玫,徐健. 类风湿关节炎患者外周血TWEAK基因启动子区甲基化状态及其表达[J]. 北京大学学报(医学版), 2021, 53(6): 1020-1025. |
| [11] | 朱忆颖,闵赛南,俞光岩. 局部注射环孢素A对非肥胖糖尿病小鼠下颌下腺分泌功能及炎症的影响[J]. 北京大学学报(医学版), 2021, 53(4): 750-757. |
| [12] | 王兆年,高文静,王碧琦,曹卫华,吕筠,余灿清,逄增昌,丛黎明,汪华,吴先萍,刘彧,李立明. 成年双生子空腹血糖、糖化血红蛋白与全基因组DNA甲基化的相关性研究[J]. 北京大学学报(医学版), 2020, 52(3): 425-431. |
| [13] | 那晓娜,朱珠,陈阳阳,王东平,王浩杰,宋阳,马晓川,王培玉,刘爱萍. 身体活动、静坐行为的时间分布与肥胖的关系[J]. 北京大学学报(医学版), 2020, 52(3): 486-491. |
| [14] | 张晓圆,郭成成,玉应香,谢岚,常翠青. 高脂饲料诱导肥胖胰岛素抵抗大鼠模型的建立[J]. 北京大学学报(医学版), 2020, 52(3): 557-563. |
| [15] | 郭成成,张晓圆,玉应香,谢岚,常翠青. 绿原酸对高脂饲料诱导的肥胖大鼠糖耐量及其曲线特征的影响[J]. 北京大学学报(医学版), 2020, 52(2): 269-274. |
|
||