北京大学学报(医学版) ›› 2021, Vol. 53 ›› Issue (1): 150-158. doi: 10.19723/j.issn.1671-167X.2021.01.023
严重急性呼吸综合征冠状病毒2的Spike蛋白点突变后与受体蛋白质及潜在抗病毒药物结合能力的同源建模分析
- 北京大学药学院天然药物及仿生药物国家重点实验室,北京 100191
Homologous modeling and binding ability analysis of Spike protein after point mutation of severe acute respiratory syndrome coronavirus 2 to receptor proteins and potential antiviral drugs
CAO Ze,WANG Le-tong,LIU Zhen-ming( )
)
- State Key Laboratory of Natural and Biomimetic Drugs, Peking University School of Pharmaceutical Sciences, Beijing 100191, China
摘要:
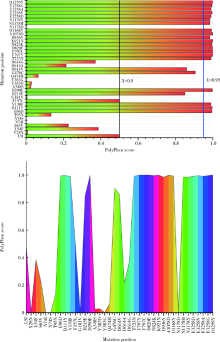
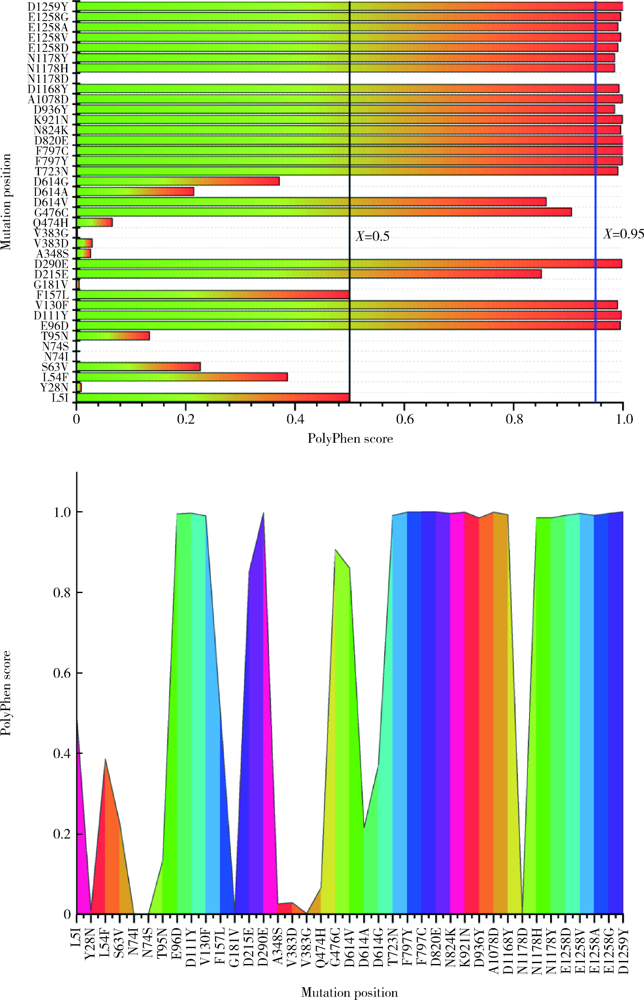
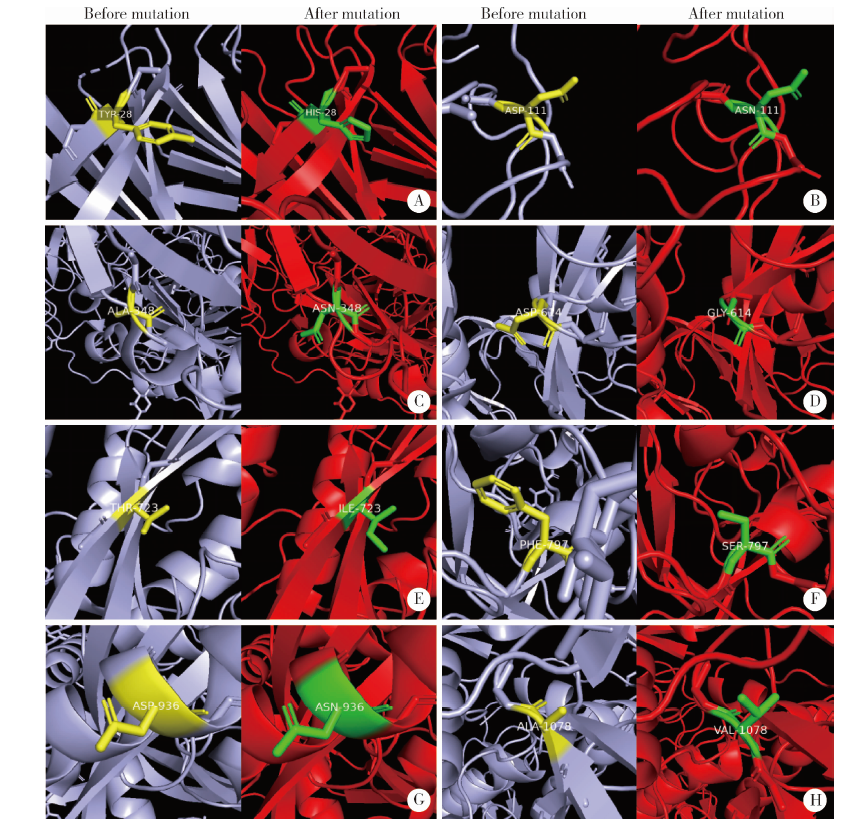
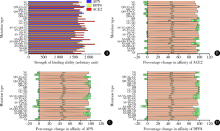
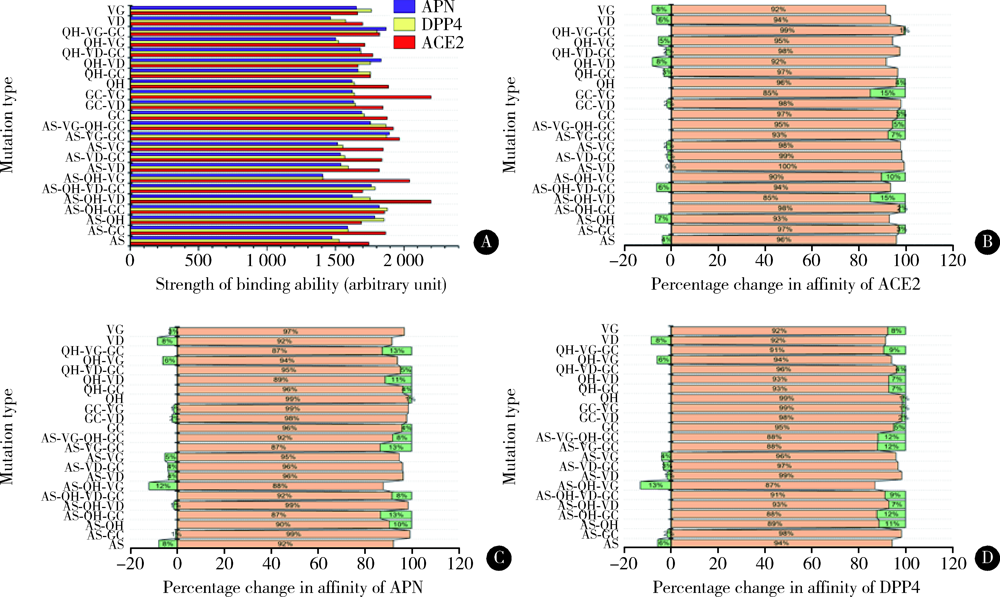
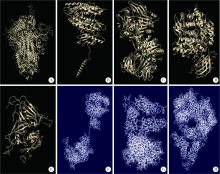
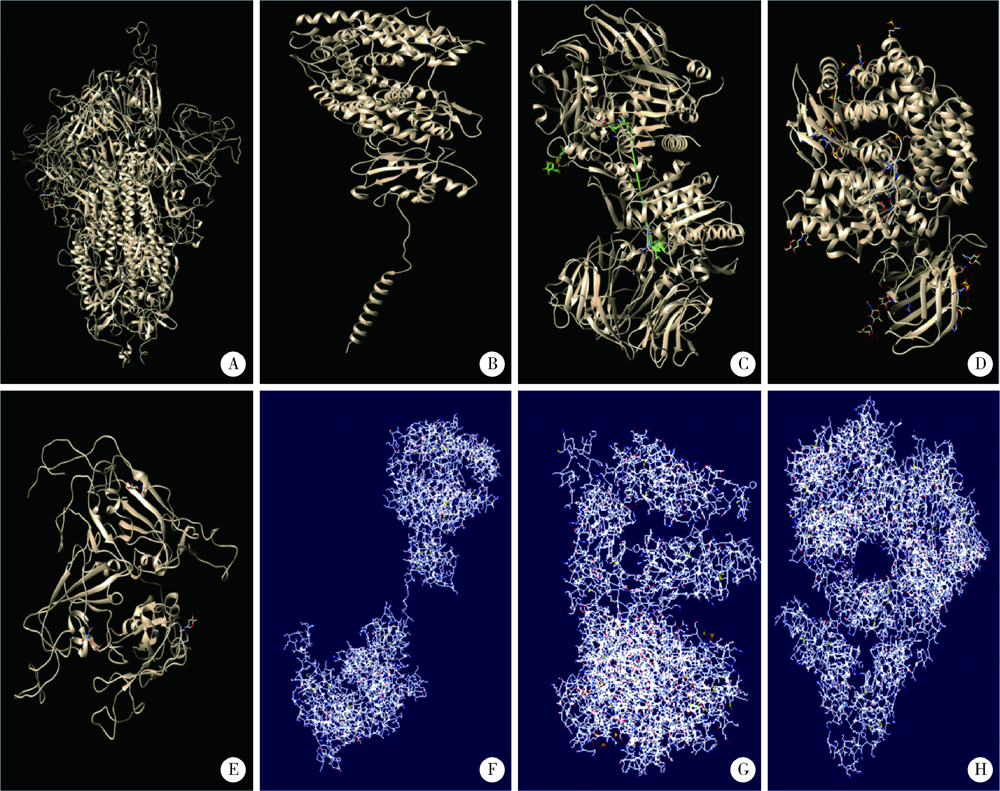
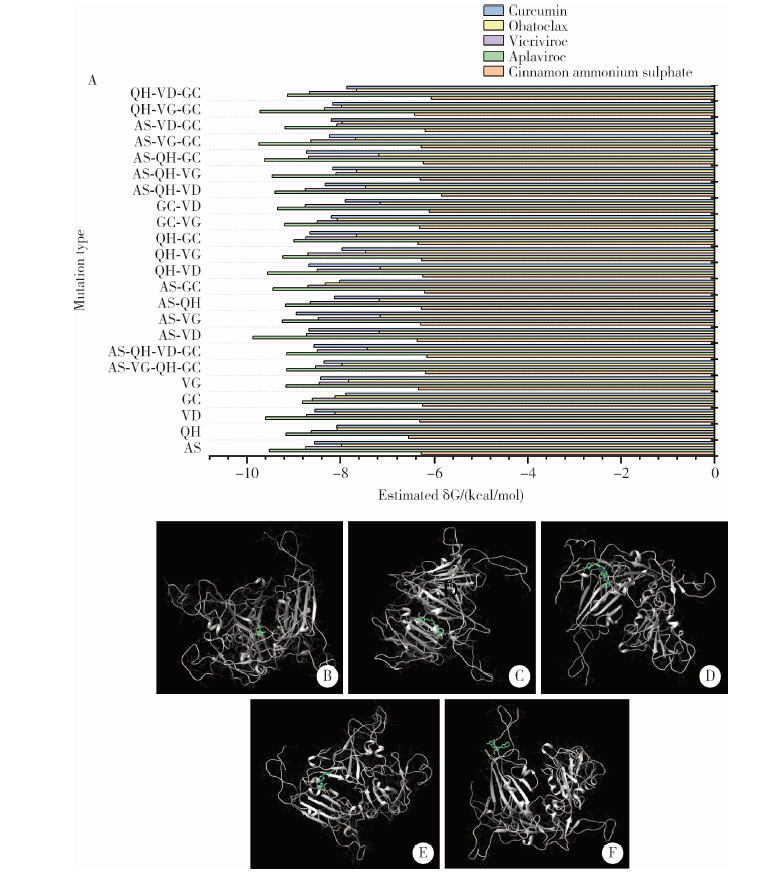
目的: 分析严重急性呼吸综合征冠状病毒2(severe acute respiratory syndrome coronavirus 2,SARS-CoV-2)的全长测序信息中,其刺突蛋白(Spike protein,S蛋白)的自发突变情况,以及S蛋白突变前后与宿主相关受体蛋白质和潜在抗病毒药物结合能力的变化。方法: 对SARS-CoV-2的一级序列进行生物信息学分析,确定高频突变位点,利用PolyPhen-2软件逐一对S蛋白突变后的功能进行预测和分析。使用SWISS-MODEL系统对突变后的S蛋白序列进行基于相似性的同源建模,利用ZDOCK程序对所建模型与血管紧张素转化酶2(angiotensin-converting enzyme 2,ACE2)、二肽基肽酶-4(dipeptidyl peptidase-4,DPP4,又称CD26)以及氨基肽酶N(aminopeptidase N,APN,又称CD13)进行蛋白质对接,用FiPD软件对结合能力评价结果进行分析,最后采用AutoDock-Chimera 1.14对突变前后的S蛋白与潜在抗病毒药物的结合能力进行预测和比较分析。结果: S蛋白的某些特定区域发生突变能够更大程度地影响其功能,突变之后,S蛋白与ACE2的结合能力趋向于减弱,而与DPP4的结合能力趋向于增强,与APN的结合能力无显著变化。抗人类免疫缺陷病毒(human immunodeficiency virus,HIV)药物aplaviroc与S蛋白的亲和能力显著高于其他候选小分子药物。结论: SARS-CoV-2在自然状态下发生突变,其S蛋白第400~1 100个氨基酸的区域为点突变高频区,突变趋势为与DPP4结合力增强,DPP4可能成为SARS-CoV-2感染细胞的新受体,aplaviroc可能成为一种SARS-CoV-2治疗药物的潜在选择。
中图分类号:
- R373.19
| [1] |
Liu J, Zheng X, Tong Q, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human patho-genic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV[J]. J Med Virol, 2020,92(5):491-494.
doi: 10.1002/jmv.25709 pmid: 32056249 |
| [2] | Wang C, Liu Z, Chen Z, et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis[J]. J Med Virol, 2020,10(1002):25762. |
| [3] |
Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex[J]. Proc Natl Acad Sci USA, 2015,112(30):9436-9441.
doi: 10.1073/pnas.1508686112 pmid: 26159422 |
| [4] |
Kim D, Lee JY, Yang JS, et al. The architecture of SARS-CoV-2 transcriptome[J]. Cell, 2020,181(4):914-921.
doi: 10.1016/j.cell.2020.04.011 pmid: 32330414 |
| [5] |
Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding[J]. Lancet, 2020,395(10224):565-574.
doi: 10.1016/S0140-6736(20)30251-8 pmid: 32007145 |
| [6] |
Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin[J]. Nature, 2020,579(7798):270-273.
doi: 10.1038/s41586-020-2012-7 pmid: 32015507 |
| [7] |
Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2[J]. Science, 2020,367(6485):1444-1448.
doi: 10.1126/science.abb2762 pmid: 32132184 |
| [8] |
Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein[J]. Cell, 2020,181(2):281-292.
doi: 10.1016/j.cell.2020.02.058 pmid: 32155444 |
| [9] |
Chan CM, Chu H, Wang Y, et al. Carcinoembryonic antigen-related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of Middle East respiratory syndrome coronavirus[J]. J Virol, 2016,90(20):9114-9127.
doi: 10.1128/JVI.01133-16 pmid: 27489282 |
| [10] |
Cui J, Li F, Shi ZL. Origin and evolution of pathogenic corona-viruses[J]. Nat Rev Microbiol, 2019,17(3):181-192.
doi: 10.1038/s41579-018-0118-9 pmid: 30531947 |
| [11] |
Chu H, Chan CM, Zhang X, et al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells[J]. J Biol Chem, 2018,293(30):11709-11726.
doi: 10.1074/jbc.RA118.001897 pmid: 29887526 |
| [12] |
Satija N, Lal SK. The molecular biology of SARS coronavirus[J]. Ann N Y Acad Sci, 2007,1102(1):26-38.
doi: 10.1196/annals.1408.002 |
| [13] |
Chen L, Gui C, Luo X, et al. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro[J]. J Virol, 2005,79(11):7095-7103.
doi: 10.1128/JVI.79.11.7095-7103.2005 pmid: 15890949 |
| [14] |
Emmelkamp JM, Rockstroh JK. CCR5 antagonists: comparison of efficacy, side effects, pharmacokinetics and interactions: review of the literature[J]. Eur J Med Res, 2007,12(9):409-417.
pmid: 17933722 |
| [15] |
Lagadinou ED, Sach A, Callahan K, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells[J]. Cell Stem Cell, 2013,12(3):329-341.
doi: 10.1016/j.stem.2012.12.013 |
| [16] |
Andersen PI, Krpina K, Ianevski A, et al. Novel antiviral activities of obatoclax, emetine, niclosamide, brequinar, and homoharringtonine[J]. Viruses, 2019,11(10):964.
doi: 10.3390/v11100964 |
| [17] |
Wen CC, Kuo YH, Jan JT, et al. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus[J]. J Med Chem, 2007,50(17):4087-4095.
doi: 10.1021/jm070295s pmid: 17663539 |
| [18] |
Aliyari Serej Z, Ebrahimi Kalan A, Mehdipour A, et al. Regulation and roles of CD26/DPPIV in hematopoiesis and diseases[J]. Biomed Pharmacother, 2017,91:88-94.
doi: 10.1016/j.biopha.2017.04.074 pmid: 28448874 |
| [19] |
Cheng F, Yuan G, He J, et al. Dysregulation of DPP4 is associa-ted with the AMPK/JAK2/STAT3 pathway in adipocytes under insulin resistance status and liraglutide intervention[J]. Diabetes Metab Syndr Obes, 2019,12:2635-2644.
doi: 10.2147/DMSO.S229838 pmid: 31849507 |
| [20] |
Song Z, Xu Y, Bao L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight[J]. Viruses, 2019,11(1):59.
doi: 10.3390/v11010059 |
| [21] |
Li Y, Zhang Z, Yang L, et al. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 Spike[J]. iScience, 2020,23(6):101160.
doi: 10.1016/j.isci.2020.101160 pmid: 32405622 |
| [22] |
Nichols WG, Steel HM, Bonny T, et al. Hepatotoxicity observed in clinical trials of aplaviroc[J]. Antimicrob Agents Chemother, 2008,52(3):858-865.
doi: 10.1128/AAC.00821-07 pmid: 18070967 |
| [23] |
Kitrinos KM, Amrine-Madsen H, Irlbeck DM, et al. Virologic failure in therapy-naive subjects on aplaviroc plus lopinavir-ritonavir: detection of aplaviroc resistance requires clonal analysis of envelope[J]. Antimicrob Agents Chemother, 2009,53(3):1124-1131.
doi: 10.1128/AAC.01057-08 pmid: 19075068 |
| [1] | 朱金荣,赵亚娜,黄巍,赵微微,王悦,王松,苏春燕. 感染新型冠状病毒的血液透析患者的临床特征[J]. 北京大学学报(医学版), 2024, 56(2): 267-272. |
| [2] | 刘鑫,石雪迎,李军. 新型冠状病毒感染相关缺血性结肠炎1例[J]. 北京大学学报(医学版), 2024, 56(2): 362-365. |
| [3] | 李建斌,吕梦娜,池强,彭一琳,刘鹏程,吴锐. 干燥综合征患者发生重症新型冠状病毒肺炎的早期预测[J]. 北京大学学报(医学版), 2023, 55(6): 1007-1012. |
| [4] | 赖金惠,王起,姬家祥,王明瑞,唐鑫伟,许克新,徐涛,胡浩. 新型冠状病毒肺炎疫情期间延迟拔除输尿管支架对泌尿系结石术后患者生活质量和心理状态的影响[J]. 北京大学学报(医学版), 2023, 55(5): 857-864. |
| [5] | 时云飞,王豪杰,刘卫平,米岚,龙孟平,刘雁飞,赖玉梅,周立新,刁新婷,李向红. 血管免疫母细胞性T细胞淋巴瘤临床与分子病理学特征分析[J]. 北京大学学报(医学版), 2023, 55(3): 521-529. |
| [6] | 熊焰,张波,聂立功,吴世凯,赵虎,李东,邸吉廷. 胸部SMARCA4缺失性未分化肿瘤的病理诊断与联合免疫检测点抑制剂治疗[J]. 北京大学学报(医学版), 2023, 55(2): 351-356. |
| [7] | 周秋君,龚潘,焦莶如,杨志仙. 1例Angelman综合征合并眼皮肤白化病2型患者的临床和遗传学分析及文献回顾[J]. 北京大学学报(医学版), 2023, 55(1): 181-185. |
| [8] | 程晓静,蒋栋,张连海,王江华,李雅真,翟佳慧,闫宝琪,张露露,谢兴旺,李子禹,季加孚. KRAS G12V特异性T细胞受体治疗恶性肿瘤的临床前研究[J]. 北京大学学报(医学版), 2022, 54(5): 884-895. |
| [9] | 尚展鹏,易阳,余蓉,范婧婧,黄昱曦,乔雪,叶敏. 荆防颗粒中抑制新型冠状病毒蛋白酶3CLpro及PLpro的活性成分[J]. 北京大学学报(医学版), 2022, 54(5): 907-919. |
| [10] | 秦彩朋,宋宇轩,丁梦婷,王飞,林佳兴,杨文博,杜依青,李清,刘士军,徐涛. 肾癌免疫治疗疗效评估突变预测模型的建立[J]. 北京大学学报(医学版), 2022, 54(4): 663-668. |
| [11] | 康志宇,王磊磊,韩永正,郭向阳. 北京冬季奥林匹克运动会运动员手术的麻醉管理[J]. 北京大学学报(医学版), 2022, 54(4): 770-773. |
| [12] | 陈曦,王斯悦,薛恩慈,王雪珩,彭和香,范梦,王梦莹,武轶群,秦雪英,李劲,吴涛,朱洪平,李静,周治波,陈大方,胡永华. 基于核心家系全外显子组测序数据探索新生突变与非综合征型唇腭裂的关联[J]. 北京大学学报(医学版), 2022, 54(3): 387-393. |
| [13] | 陈明隆,刘笑晗,郭静. 新型冠状病毒肺炎疫情下儿童父母社会支持与养育倦怠的关系[J]. 北京大学学报(医学版), 2022, 54(3): 520-525. |
| [14] | 杨林承,张瑞涛,郭丽君,肖晗,祖凌云,张幼怡,程秦,赵志伶,葛庆岗,高炜. 低氧状态及炎症反应是新型冠状病毒肺炎患者发生急性心肌损伤的危险因素[J]. 北京大学学报(医学版), 2021, 53(1): 159-166. |
| [15] | 吴君怡,余淼,孙仕晨,樊壮壮,郑静蕾,张刘陶,冯海兰,刘洋,韩冬. 少汗性外胚层发育不良患者EDA基因突变检测及表型分析[J]. 北京大学学报(医学版), 2021, 53(1): 24-33. |
|
||